A rapidly evolving genomic toolkit for Drosophila heterochromatin
- PMID: 23519206
- PMCID: PMC4049844
- DOI: 10.4161/fly.24335
A rapidly evolving genomic toolkit for Drosophila heterochromatin
Abstract
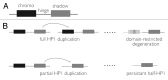
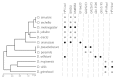
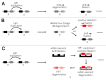
Heterochromatin is the enigmatic eukaryotic genome compartment found mostly at telomeres and centromeres. Conventional approaches to sequence assembly and genetic manipulation fail in this highly repetitive, gene-sparse, and recombinationally silent DNA. In contrast, genetic and molecular analyses of euchromatin-encoded proteins that bind, remodel, and propagate heterochromatin have revealed its vital role in numerous cellular and evolutionary processes. Utilizing the 12 sequenced Drosophila genomes, Levine et al (1) took a phylogenomic approach to discover new such protein "surrogates" of heterochromatin function and evolution. This paper reported over 20 new members of what was traditionally believed to be a small and static Heterochromatin Protein 1 (HP1) gene family. The newly identified HP1 proteins are structurally diverse, lineage-restricted, and expressed primarily in the male germline. The birth and death of HP1 genes follows a "revolving door" pattern, where new HP1s appear to replace old HP1s. Here, we address alternative evolutionary models that drive this constant innovation.
Keywords: Drosophila; HP1; Heterochromatin Protein 1; chromodomain; chromoshadow domain; gene duplication; germline; heterochromatin; phylogenomics; pseudogenization.
Figures
Comment on
-
Phylogenomic analysis reveals dynamic evolutionary history of the Drosophila heterochromatin protein 1 (HP1) gene family.PLoS Genet. 2012;8(6):e1002729. doi: 10.1371/journal.pgen.1002729. Epub 2012 Jun 21. PLoS Genet. 2012. PMID: 22737079 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
