Atomic structure of the 75 MDa extremophile Sulfolobus turreted icosahedral virus determined by CryoEM and X-ray crystallography
- PMID: 23520050
- PMCID: PMC3619359
- DOI: 10.1073/pnas.1300601110
Atomic structure of the 75 MDa extremophile Sulfolobus turreted icosahedral virus determined by CryoEM and X-ray crystallography
Abstract
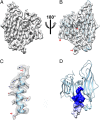
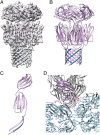
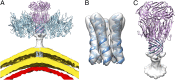
Sulfolobus turreted icosahedral virus (STIV) was isolated in acidic hot springs where it infects the archeon Sulfolobus solfataricus. We determined the STIV structure using near-atomic resolution electron microscopy and X-ray crystallography allowing tracing of structural polypeptide chains and visualization of transmembrane proteins embedded in the viral membrane. We propose that the vertex complexes orchestrate virion assembly by coordinating interactions of the membrane and various protein components involved. STIV shares the same coat subunit and penton base protein folds as some eukaryotic and bacterial viruses, suggesting that they derive from a common ancestor predating the divergence of the three kingdoms of life. One architectural motif (β-jelly roll fold) forms virtually the entire capsid (distributed in three different gene products), indicating that a single ancestral protein module may have been at the origin of its evolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

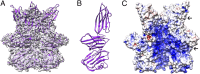

Similar articles
-
The architecture and chemical stability of the archaeal Sulfolobus turreted icosahedral virus.J Virol. 2010 Sep;84(18):9575-83. doi: 10.1128/JVI.00708-10. Epub 2010 Jun 30. J Virol. 2010. PMID: 20592081 Free PMC article.
-
Structure-Based Mutagenesis of Sulfolobus Turreted Icosahedral Virus B204 Reveals Essential Residues in the Virion-Associated DNA-Packaging ATPase.J Virol. 2015 Dec 23;90(6):2729-39. doi: 10.1128/JVI.02435-15. J Virol. 2015. PMID: 26699645 Free PMC article.
-
Characterization of the archaeal thermophile Sulfolobus turreted icosahedral virus validates an evolutionary link among double-stranded DNA viruses from all domains of life.J Virol. 2006 Aug;80(15):7625-35. doi: 10.1128/JVI.00522-06. J Virol. 2006. PMID: 16840341 Free PMC article.
-
Structure and cell biology of archaeal virus STIV.Curr Opin Virol. 2012 Apr;2(2):122-7. doi: 10.1016/j.coviro.2012.01.007. Epub 2012 Mar 20. Curr Opin Virol. 2012. PMID: 22482708 Free PMC article. Review.
-
Genetics, biochemistry and structure of the archaeal virus STIV.Biochem Soc Trans. 2009 Feb;37(Pt 1):114-7. doi: 10.1042/BST0370114. Biochem Soc Trans. 2009. PMID: 19143613 Review.
Cited by
-
Quantification of Protein-Induced Membrane Remodeling Kinetics In Vitro with Lipid Multilayer Gratings.Small. 2016 Jan 27;12(4):506-15. doi: 10.1002/smll.201502398. Epub 2015 Dec 9. Small. 2016. PMID: 26649649 Free PMC article.
-
Unique architecture of thermophilic archaeal virus APBV1 and its genome packaging.Nat Commun. 2017 Nov 10;8(1):1436. doi: 10.1038/s41467-017-01668-0. Nat Commun. 2017. PMID: 29127347 Free PMC article.
-
An extensively glycosylated archaeal pilus survives extreme conditions.Nat Microbiol. 2019 Aug;4(8):1401-1410. doi: 10.1038/s41564-019-0458-x. Epub 2019 May 20. Nat Microbiol. 2019. PMID: 31110358 Free PMC article.
-
Structural basis for assembly of vertical single β-barrel viruses.Nat Commun. 2019 Mar 12;10(1):1184. doi: 10.1038/s41467-019-08927-2. Nat Commun. 2019. PMID: 30862777 Free PMC article.
-
Bipartite Network Analysis of the Archaeal Virosphere: Evolutionary Connections between Viruses and Capsidless Mobile Elements.J Virol. 2016 Nov 28;90(24):11043-11055. doi: 10.1128/JVI.01622-16. Print 2016 Dec 15. J Virol. 2016. PMID: 27681128 Free PMC article.
References
-
- Prangishvili D, Forterre P, Garrett RA. Viruses of the Archaea: A unifying view. Nat Rev Microbiol. 2006;4(11):837–848. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

