Metabolic adaptation to chronic inhibition of mitochondrial protein synthesis in acute myeloid leukemia cells
- PMID: 23520503
- PMCID: PMC3592803
- DOI: 10.1371/journal.pone.0058367
Metabolic adaptation to chronic inhibition of mitochondrial protein synthesis in acute myeloid leukemia cells
Abstract
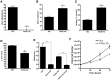
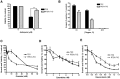
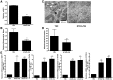
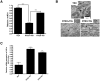
Recently, we demonstrated that the anti-bacterial agent tigecycline preferentially induces death in leukemia cells through the inhibition of mitochondrial protein synthesis. Here, we sought to understand mechanisms of resistance to tigecycline by establishing a leukemia cell line resistant to the drug. TEX leukemia cells were treated with increasing concentrations of tigecycline over 4 months and a population of cells resistant to tigecycline (RTEX+TIG) was selected. Compared to wild type cells, RTEX+TIG cells had undetectable levels of mitochondrially translated proteins Cox-1 and Cox-2, reduced oxygen consumption and increased rates of glycolysis. Moreover, RTEX+TIG cells were more sensitive to inhibitors of glycolysis and more resistant to hypoxia. By electron microscopy, RTEX+TIG cells had abnormally swollen mitochondria with irregular cristae structures. RNA sequencing demonstrated a significant over-representation of genes with binding sites for the HIF1α:HIF1β transcription factor complex in their promoters. Upregulation of HIF1α mRNA and protein in RTEX+TIG cells was confirmed by Q-RTPCR and immunoblotting. Strikingly, upon removal of tigecycline from RTEX+TIG cells, the cells re-established aerobic metabolism. Levels of Cox-1 and Cox-2, oxygen consumption, glycolysis, mitochondrial mass and mitochondrial membrane potential returned to wild type levels, but HIF1α remained elevated. However, upon re-treatment with tigecycline for 72 hours, the glycolytic phenotype was re-established. Thus, we have generated cells with a reversible metabolic phenotype by chronic treatment with an inhibitor of mitochondrial protein synthesis. These cells will provide insight into cellular adaptations used to cope with metabolic stress.
Conflict of interest statement
Figures




References
-
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, et al. (1981) Sequence and organization of the human mitochondrial genome. Nature 290: 457–465. - PubMed
-
- King MP, Attardi G (1989) Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science 246: 500–503. - PubMed
-
- Inoue K, Takai D, Hosaka H, Ito S, Shitara H, et al. (1997) Isolation and characterization of mitochondrial DNA-less lines from various mammalian cell lines by application of an anticancer drug, ditercalinium. Biochem Biophys Res Commun 239: 257–260. - PubMed
-
- Warner JK, Wang JC, Takenaka K, Doulatov S, McKenzie JL, et al. (2005) Direct evidence for cooperating genetic events in the leukemic transformation of normal human hematopoietic cells. Leukemia 19: 1794–1805. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

