Derepression of Cancer/testis antigens in cancer is associated with distinct patterns of DNA hypomethylation
- PMID: 23522060
- PMCID: PMC3618251
- DOI: 10.1186/1471-2407-13-144
Derepression of Cancer/testis antigens in cancer is associated with distinct patterns of DNA hypomethylation
Abstract
Background: The Cancer/Testis Antigens (CTAs) are a heterogeneous group of proteins whose expression is typically restricted to the testis. However, they are aberrantly expressed in most cancers that have been examined to date. Broadly speaking, the CTAs can be divided into two groups: the CTX antigens that are encoded by the X-linked genes and the non-X CT antigens that are encoded by the autosomes. Unlike the non-X CTAs, the CTX antigens form clusters of closely related gene families and their expression is frequently associated with advanced disease with poorer prognosis. Regardless however, the mechanism(s) underlying their selective derepression and stage-specific expression in cancer remain poorly understood, although promoter DNA demethylation is believed to be the major driver.
Methods: Here, we report a systematic analysis of DNA methylation profiling data from various tissue types to elucidate the mechanism underlying the derepression of the CTAs in cancer. We analyzed the methylation profiles of 501 samples including sperm, several cancer types, and their corresponding normal somatic tissue types.
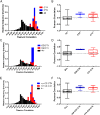
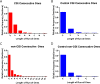
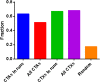
Results: We found strong evidence for specific DNA hypomethylation of CTA promoters in the testis and cancer cells but not in their normal somatic counterparts. We also found that hypomethylation was clustered on the genome into domains that coincided with nuclear lamina-associated domains (LADs) and that these regions appeared to be insulated by CTCF sites. Interestingly, we did not observe any significant differences in the hypomethylation pattern between the CTAs without CpG islands and the CTAs with CpG islands in the proximal promoter.
Conclusion: Our results corroborate that widespread DNA hypomethylation appears to be the driver in the derepression of CTA expression in cancer and furthermore, demonstrate that these hypomethylated domains are associated with the nuclear lamina-associated domains (LADS). Taken together, our results suggest that wide-spread methylation changes in cancer are linked to derepression of germ-line-specific genes that is orchestrated by the three dimensional organization of the cancer genome.
Figures


Similar articles
-
Reciprocal binding of CTCF and BORIS to the NY-ESO-1 promoter coincides with derepression of this cancer-testis gene in lung cancer cells.Cancer Res. 2005 Sep 1;65(17):7763-74. doi: 10.1158/0008-5472.CAN-05-0823. Cancer Res. 2005. PMID: 16140944
-
Conditional expression of the CTCF-paralogous transcriptional factor BORIS in normal cells results in demethylation and derepression of MAGE-A1 and reactivation of other cancer-testis genes.Cancer Res. 2005 Sep 1;65(17):7751-62. doi: 10.1158/0008-5472.CAN-05-0858. Cancer Res. 2005. PMID: 16140943
-
A new high-throughput screening methodology for the discovery of cancer-testis antigen using multi-omics data.Comput Methods Programs Biomed. 2024 Jun;250:108193. doi: 10.1016/j.cmpb.2024.108193. Epub 2024 Apr 25. Comput Methods Programs Biomed. 2024. PMID: 38678957
-
Cancer-testis antigens: Unique cancer stem cell biomarkers and targets for cancer therapy.Semin Cancer Biol. 2018 Dec;53:75-89. doi: 10.1016/j.semcancer.2018.08.006. Epub 2018 Aug 29. Semin Cancer Biol. 2018. PMID: 30171980 Review.
-
Cancer/Testis Antigens: Expression, Regulation, Tumor Invasion, and Use in Immunotherapy of Cancers.Immunol Invest. 2016 Oct;45(7):619-40. doi: 10.1080/08820139.2016.1197241. Epub 2016 Sep 7. Immunol Invest. 2016. PMID: 27603913 Review.
Cited by
-
Profiling cancer testis antigens in non-small-cell lung cancer.JCI Insight. 2016 Jul 7;1(10):e86837. doi: 10.1172/jci.insight.86837. JCI Insight. 2016. PMID: 27699219 Free PMC article.
-
Expression of Cancer Testis Antigens in Colorectal Cancer: New Prognostic and Therapeutic Implications.Dis Markers. 2016;2016:1987505. doi: 10.1155/2016/1987505. Epub 2016 Aug 18. Dis Markers. 2016. PMID: 27635108 Free PMC article.
-
Fundamental diversity of human CpG islands at multiple biological levels.Epigenetics. 2014 Apr;9(4):483-91. doi: 10.4161/epi.27654. Epub 2014 Jan 13. Epigenetics. 2014. PMID: 24419148 Free PMC article.
-
Cancer testis antigens and genomic instability: More than immunology.DNA Repair (Amst). 2021 Dec;108:103214. doi: 10.1016/j.dnarep.2021.103214. Epub 2021 Aug 17. DNA Repair (Amst). 2021. PMID: 34481156 Free PMC article. Review.
-
Cancer testis antigens: Emerging therapeutic targets leveraging genomic instability in cancer.Mol Ther Oncol. 2024 Jan 26;32(1):200768. doi: 10.1016/j.omton.2024.200768. eCollection 2024 Mar 21. Mol Ther Oncol. 2024. PMID: 38596293 Free PMC article. Review.
References
-
- Scanlan MJ, Simpson AJ, Old LJ. The cancer/testis genes: review, standardization, and commentary. Cancer Immun. 2004;4:1. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

