Non-negative matrix factorization by maximizing correntropy for cancer clustering
- PMID: 23522344
- PMCID: PMC3659102
- DOI: 10.1186/1471-2105-14-107
Non-negative matrix factorization by maximizing correntropy for cancer clustering
Abstract
Background: Non-negative matrix factorization (NMF) has been shown to be a powerful tool for clustering gene expression data, which are widely used to classify cancers. NMF aims to find two non-negative matrices whose product closely approximates the original matrix. Traditional NMF methods minimize either the l2 norm or the Kullback-Leibler distance between the product of the two matrices and the original matrix. Correntropy was recently shown to be an effective similarity measurement due to its stability to outliers or noise.
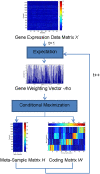
Results: We propose a maximum correntropy criterion (MCC)-based NMF method (NMF-MCC) for gene expression data-based cancer clustering. Instead of minimizing the l2 norm or the Kullback-Leibler distance, NMF-MCC maximizes the correntropy between the product of the two matrices and the original matrix. The optimization problem can be solved by an expectation conditional maximization algorithm.
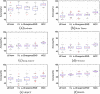
Conclusions: Extensive experiments on six cancer benchmark sets demonstrate that the proposed method is significantly more accurate than the state-of-the-art methods in cancer clustering.
Figures




References
-
- Gao Y, Church G. Improving molecular cancer class discovery through sparse non-negative matrix factorization. Bioinformatics. 2005;21(21):3970—3975. - PubMed
-
- Zheng CH, Ng TY, Zhang L, Shiu CK, Wang HQ. Tumor classification based on non-negative matrix factorization using gene expression data. IEEE Trans Nanobioscience. 2011;10(2):86–93. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

