Evidence for monomeric actin function in INO80 chromatin remodeling
- PMID: 23524535
- PMCID: PMC3618487
- DOI: 10.1038/nsmb.2529
Evidence for monomeric actin function in INO80 chromatin remodeling
Abstract
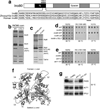
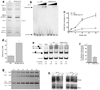
Actin has well-established functions in the cytoplasm, but its roles in the nucleus remain poorly defined. Here, by studying the nuclear actin-containing yeast INO80 chromatin remodeling complex, we provide genetic and biochemical evidence for a role of monomeric actin in INO80 chromatin remodeling. We demonstrate that, in contrast to cytoplasmic actin, nuclear actin is present as a monomer in the INO80 complex, and its barbed end is not accessible for polymerization. We identify an actin mutation in subdomain 2 affecting in vivo nuclear functions and reducing the chromatin remodeling activity of the INO80 complex in vitro. Notably, the highly conserved subdomain 2 at the pointed end of actin contributes to the interaction of INO80 with chromatin. Our results establish an evolutionarily conserved function of nuclear actin in its monomeric form and suggest that nuclear actin can utilize a fundamentally distinct mechanism from that of cytoplasmic actin.
Figures



Comment in
-
Monomeric actin required for INO80 remodeling.Nat Struct Mol Biol. 2013 Apr;20(4):405-7. doi: 10.1038/nsmb.2553. Nat Struct Mol Biol. 2013. PMID: 23552289 No abstract available.
References
-
- Bettinger BT, Gilbert DM, Amberg DC. Actin up in the nucleus. Nat Rev Mol Cell Biol. 2004;5:410–415. - PubMed
-
- Blessing CA, Ugrinova GT, Goodson HV. Actin and ARPs: action in the nucleus. Trends Cell Biol. 2004;14:435–442. - PubMed
-
- Olave IA, Reck-Peterson SL, Crabtree GR. Nuclear actin and actin-related proteins in chromatin remodeling. Annu Rev Biochem. 2002;71:755–781. - PubMed
-
- Shumaker DK, Kuczmarski ER, Goldman RD. The nucleoskeleton: lamins and actin are major players in essential nuclear functions. Curr Opin Cell Biol. 2003;15:358–366. - PubMed
-
- Clark TG, Merriam RW. Diffusible and bound actin nuclei of Xenopus laevis oocytes. Cell. 1977;12:883–891. - PubMed
Methods-only references
-
- Shen X. Preparation and analysis of the INO80 complex. Methods Enzymol. 2004;377:401–412. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

