TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h
- PMID: 23524850
- PMCID: PMC3630359
- DOI: 10.1038/emboj.2013.61
TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h
Abstract
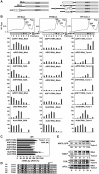
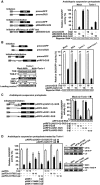
Mammalian target-of-rapamycin (mTOR) triggers S6 kinase (S6K) activation to phosphorylate targets linked to translation in response to energy, nutrients, and hormones. Pathways of TOR activation in plants remain unknown. Here, we uncover the role of the phytohormone auxin in TOR signalling activation and reinitiation after upstream open reading frame (uORF) translation, which in plants is dependent on translation initiation factor eIF3h. We show that auxin triggers TOR activation followed by S6K1 phosphorylation at T449 and efficient loading of uORF-mRNAs onto polysomes in a manner sensitive to the TOR inhibitor Torin-1. Torin-1 mediates recruitment of inactive S6K1 to polysomes, while auxin triggers S6K1 dissociation and recruitment of activated TOR instead. A putative target of TOR/S6K1-eIF3h-is phosphorylated and detected in polysomes in response to auxin. In TOR-deficient plants, polysomes were prebound by inactive S6K1, and loading of uORF-mRNAs and eIF3h was impaired. Transient expression of eIF3h-S178D in plant protoplasts specifically upregulates uORF-mRNA translation. We propose that TOR functions in polysomes to maintain the active S6K1 (and thus eIF3h) phosphorylation status that is critical for translation reinitiation.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures







Comment in
-
TOR tour to auxin.EMBO J. 2013 Apr 17;32(8):1069-71. doi: 10.1038/emboj.2013.69. Epub 2013 Mar 22. EMBO J. 2013. PMID: 23524852 Free PMC article.
References
-
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu W-L, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. EMBO J 415: 977–983 - PubMed
-
- Beltrán-Peña E, Aguilar R, Ortíz-López A, Dinkova TD, De Jiménez ES (2002) Auxin stimulates S6 ribosomal protein phosphorylation in maize thereby affecting protein synthesis regulation. Physiol Plant 115: 291–297 - PubMed
-
- Browning KS, Gallie DR, Hershey JWB, Hinnebusch AG, Maitra U, Merrick WC, Norbury C (2001) Unified nomenclature for the subunits of eukaryotic initiation factor 3. Trends Biochem Sci 26: 284. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

