SENP3-mediated deSUMOylation of dynamin-related protein 1 promotes cell death following ischaemia
- PMID: 23524851
- PMCID: PMC3671254
- DOI: 10.1038/emboj.2013.65
SENP3-mediated deSUMOylation of dynamin-related protein 1 promotes cell death following ischaemia
Abstract
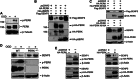
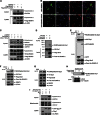
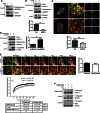
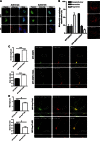
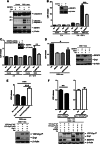
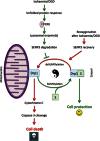
Global increases in small ubiquitin-like modifier (SUMO)-2/3 conjugation are a neuroprotective response to severe stress but the mechanisms and specific target proteins that determine cell survival have not been identified. Here, we demonstrate that the SUMO-2/3-specific protease SENP3 is degraded during oxygen/glucose deprivation (OGD), an in vitro model of ischaemia, via a pathway involving the unfolded protein response (UPR) kinase PERK and the lysosomal enzyme cathepsin B. A key target for SENP3-mediated deSUMOylation is the GTPase Drp1, which plays a major role in regulating mitochondrial fission. We show that depletion of SENP3 prolongs Drp1 SUMOylation, which suppresses Drp1-mediated cytochrome c release and caspase-mediated cell death. SENP3 levels recover following reoxygenation after OGD allowing deSUMOylation of Drp1, which facilitates Drp1 localization at mitochondria and promotes fragmentation and cytochrome c release. RNAi knockdown of SENP3 protects cells from reoxygenation-induced cell death via a mechanism that requires Drp1 SUMOylation. Thus, we identify a novel adaptive pathway to extreme cell stress in which dynamic changes in SENP3 stability and regulation of Drp1 SUMOylation are crucial determinants of cell fate.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

Comment in
-
Post-translational modification: A SUMO protease for stress protection.Nat Rev Mol Cell Biol. 2013 May;14(5):263. doi: 10.1038/nrm3569. Epub 2013 Apr 18. Nat Rev Mol Cell Biol. 2013. PMID: 23594955 No abstract available.
-
SUMO wrestling with Drp1 at mitochondria.EMBO J. 2013 May 29;32(11):1496-8. doi: 10.1038/emboj.2013.103. Epub 2013 Apr 30. EMBO J. 2013. PMID: 23632859 Free PMC article.
References
-
- Boya P, Kroemer G (2008) Lysosomal membrane permeabilization in cell death. Oncogene 27: 6434–6451 - PubMed
-
- Cassidy-Stone A, Chipuk JE, Ingerman E, Song C, Yoo C, Kuwana T, Kurth MJ, Shaw JT, Hinshaw JE, Green DR, Nunnari J (2008) Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev Cell 14: 193–204 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

