A library of programmable DNAzymes that operate in a cellular environment
- PMID: 23525068
- PMCID: PMC3607177
- DOI: 10.1038/srep01535
A library of programmable DNAzymes that operate in a cellular environment
Abstract
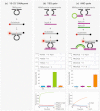
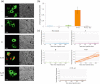
DNAzymes were used as inhibitory agents in a variety of experimental disease settings, such as cancer, viral infections and even HIV. Drugs that become active only upon the presence of preprogrammed abnormal environmental conditions may enable selective molecular therapy by targeting abnormal cells without injuring normal cells. Here we show a novel programmable DNAzyme library composed of variety of Boolean logic gates, including YES, AND, NOT, OR, NAND, ANDNOT, XOR, NOR and 3-input-AND gate, that uses both miRNAs and mRNAs as inputs. Each gate is based on the c-jun cleaving Dz13 DNAzyme and active only in the presence of specific input combinations. The library is modular, supports arbitrary inputs and outputs, cascadable, highly specific and robust. We demonstrate the library's potential diagnostic abilities on miRNA and mRNA combinations in cell lysate and its ability to operate in a cellular environment by using beacon-like c-jun mimicking substrate in living mammalian cells.
Figures


Similar articles
-
The DNAzymes Rs6, Dz13, and DzF have potent biologic effects independent of catalytic activity.Oligonucleotides. 2006 Winter;16(4):297-312. doi: 10.1089/oli.2006.16.297. Oligonucleotides. 2006. PMID: 17155906
-
DNA computing circuits using libraries of DNAzyme subunits.Nat Nanotechnol. 2010 Jun;5(6):417-22. doi: 10.1038/nnano.2010.88. Epub 2010 May 30. Nat Nanotechnol. 2010. PMID: 20512129
-
Towards the development of a DNA automaton: modular RNA-cleaving deoxyribozyme logic gates regulated by miRNAs.Analyst. 2024 Mar 11;149(6):1947-1957. doi: 10.1039/d3an02178e. Analyst. 2024. PMID: 38385166
-
Dz13: c-Jun downregulation and tumour cell death.Chem Biol Drug Des. 2011 Dec;78(6):909-12. doi: 10.1111/j.1747-0285.2011.01166.x. Epub 2011 Oct 31. Chem Biol Drug Des. 2011. PMID: 21722318 Review.
-
Theranostic DNAzymes.Theranostics. 2017 Feb 23;7(4):1010-1025. doi: 10.7150/thno.17736. eCollection 2017. Theranostics. 2017. PMID: 28382172 Free PMC article. Review.
Cited by
-
Continuous variables logic via coupled automata using a DNAzyme cascade with feedback.Chem Sci. 2017 Mar 1;8(3):2161-2168. doi: 10.1039/c6sc03892a. Epub 2016 Dec 1. Chem Sci. 2017. PMID: 28507669 Free PMC article.
-
Design of a biochemical circuit motif for learning linear functions.J R Soc Interface. 2014 Dec 6;11(101):20140902. doi: 10.1098/rsif.2014.0902. J R Soc Interface. 2014. PMID: 25401175 Free PMC article.
-
Biosensors with built-in biomolecular logic gates for practical applications.Biosensors (Basel). 2014 Aug 27;4(3):273-300. doi: 10.3390/bios4030273. eCollection 2014 Sep. Biosensors (Basel). 2014. PMID: 25587423 Free PMC article. Review.
-
Signal propagation in multi-layer DNAzyme cascades using structured chimeric substrates.Angew Chem Int Ed Engl. 2014 Jul 7;53(28):7183-7. doi: 10.1002/anie.201402691. Epub 2014 Jun 2. Angew Chem Int Ed Engl. 2014. PMID: 24890874 Free PMC article.
-
Melanoma protective antitumor immunity activated by catalytic DNA.Oncogene. 2018 Sep;37(37):5115-5126. doi: 10.1038/s41388-018-0306-0. Epub 2018 May 29. Oncogene. 2018. PMID: 29844573
References
-
- Breaker R. R. & Joyce G. F. A DNA enzyme that cleaves RNA. Chem Biol 1, 223–229 (1994). - PubMed
-
- Dass C. R., Galloway S. J. & Choong P. F. Dz13, a c-jun DNAzyme, is a potent inducer of caspase-2 activation. Oligonucleotides 20, 137–146 (2010). - PubMed
-
- Wu Y. et al. Inhibition of bcr-abl oncogene expression by novel deoxyribozymes (DNAzymes). Hum Gene Ther 10, 2847–2857 (1999). - PubMed
-
- Dass C. R., Choong P. F. & Khachigian L. M. DNAzyme technology and cancer therapy: cleave and let die. Mol Cancer Ther 7, 243–251 (2008). - PubMed
-
- Reyes-Gutiérrez P. & Alvarez-Salas L. M. Cleavage of HPV-16 E6/E7 mRNA mediated by modified 10-23 deoxyribozymes. Oligonucleotides 19, 233–242 (2009). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

