Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation
- PMID: 23525088
- PMCID: PMC3672957
- DOI: 10.1038/ni.2556
Control of amino-acid transport by antigen receptors coordinates the metabolic reprogramming essential for T cell differentiation
Erratum in
- Nat Immunol. 2014 Jan;15(1):109
Abstract
T lymphocytes must regulate nutrient uptake to meet the metabolic demands of an immune response. Here we show that the intracellular supply of large neutral amino acids (LNAAs) in T cells was regulated by pathogens and the T cell antigen receptor (TCR). T cells responded to antigen by upregulating expression of many amino-acid transporters, but a single System L ('leucine-preferring system') transporter, Slc7a5, mediated uptake of LNAAs in activated T cells. Slc7a5-null T cells were unable to metabolically reprogram in response to antigen and did not undergo clonal expansion or effector differentiation. The metabolic catastrophe caused by loss of Slc7a5 reflected the requirement for sustained uptake of the LNAA leucine for activation of the serine-threonine kinase complex mTORC1 and for expression of the transcription factor c-Myc. Control of expression of the System L transporter by pathogens is thus a critical metabolic checkpoint for T cells.
Figures






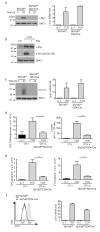
Comment in
-
Slc7a5 helps T cells get with the program.Nat Immunol. 2013 May;14(5):422-4. doi: 10.1038/ni.2594. Nat Immunol. 2013. PMID: 23598390 No abstract available.
References
-
- Wang R, Green DR. Metabolic checkpoints in activated T cells. Nat. Immunol. 2012;13:907–915. - PubMed
-
- Schriever SC, Deutsch MJ, Adamski J, Roscher AA, Ensenauer R. Cellular signaling of amino acids towards mTORC1 activation in impaired human leucine catabolism. The Journal of Nutritional Biochemistry. 2012 doi:10.1016/j.jnutbio.2012.04.018. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

