Hysteresis as a Marker for Complex, Overlapping Landscapes in Proteins
- PMID: 23525263
- PMCID: PMC3601837
- DOI: 10.1021/jz301893w
Hysteresis as a Marker for Complex, Overlapping Landscapes in Proteins
Abstract
Topologically complex proteins fold by multiple routes as a result of hard-to-fold regions of the proteins. Oftentimes these regions are introduced into the protein scaffold for function and increase frustration in the otherwise smooth-funneled landscape. Interestingly, while functional regions add complexity to folding landscapes, they may also contribute to a unique behavior referred to as hysteresis. While hysteresis is predicted to be rare, it is observed in various proteins, including proteins containing a unique peptide cyclization to form a fluorescent chromophore as well as proteins containing a knotted topology in their native fold. Here, hysteresis is demonstrated to be a consequence of the decoupling of unfolding events from the isomerization or hula-twist of a chromophore in one protein and the untying of the knot in a second protein system. The question now is- can hysteresis be a marker for the interplay of landscapes where complex folding and functional regions overlap?
Keywords: Energy Landscape; Interplay; Knotted Proteins; Protein Folding.
Figures
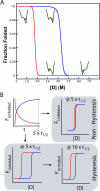
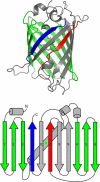


References
-
- Levinthal C. Are There Pathways for Protein Folding? Extrait du Journal De Chimie Physique. 1968;65(1):44–7.
-
- Weissman JS. All roads lead to Rome? The multiple pathways of protein folding. Chem Biol. 1995;2(5):255–60. - PubMed
-
- Dill KA, Chan HS. From Levinthal to pathways to funnels. Nature structural biology. 1997;4(1):10–9. - PubMed
-
- Onuchic JN, Luthey-Schulten Z, Wolynes PG. Theory of protein folding: the energy landscape perspective. Annu Rev Phys Chem. 1997;48:545–600. - PubMed
-
- Onuchic JN, Wolynes PG. Theory of protein folding. Current opinion in structural biology. 2004;14(1):70–5. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

