A novel bifunctional histone protein in Streptomyces: a candidate for structural coupling between DNA conformation and transcription during development and stress?
- PMID: 23525459
- PMCID: PMC3643593
- DOI: 10.1093/nar/gkt180
A novel bifunctional histone protein in Streptomyces: a candidate for structural coupling between DNA conformation and transcription during development and stress?
Abstract
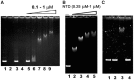
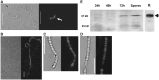
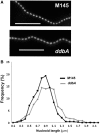
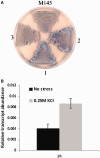
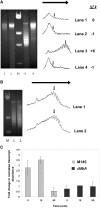
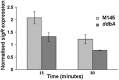
Antibiotic-producing Streptomyces are complex bacteria that remodel global transcription patterns and their nucleoids during development. Here, we describe a novel developmentally regulated nucleoid-associated protein, DdbA, of the genus that consists of an N-terminal DNA-binding histone H1-like domain and a C-terminal DksA-like domain that can potentially modulate RNA polymerase activity in conjunction with ppGpp. Owing to its N-terminal domain, the protein can efficiently bind and condense DNA in vitro. Loss of function of this DNA-binding protein results in changes in both DNA condensation during development and the ability to adjust DNA supercoiling in response to osmotic stress. Initial analysis of the DksA-like activity of DdbA indicates that overexpression of the protein suppresses a conditional deficiency in antibiotic production of relA mutants that are unable to synthesise ppGpp, just as DksA overexpression in Escherichia coli can suppress ppGpp(0) phenotypes. The null mutant is also sensitive to oxidative stress owing to impaired upregulation of transcription of sigR, encoding an alternative sigma factor. Consequently, we propose this bifunctional histone-like protein as a candidate that could structurally couple changes in DNA conformation and transcription during the streptomycete life-cycle and in response to stress.
Figures


References
-
- Martin JF, Liras P. Engineering of regulatory cascades and networks controlling antibiotic biosynthesis in Streptomyces. Curr. Opin. Microbiol. 13:263–273. - PubMed
-
- Hesketh A, Sun J, Bibb M. Induction of ppGpp synthesis in Streptomyces coelicolor A3(2) grown under conditions of nutritional sufficiency elicits actII-ORF4 transcription and actinorhodin biosynthesis. Mol. Microbiol. 2001;39:136–144. - PubMed
-
- Potrykus K, Cashel M. (p)ppGpp: still magical? Annu. Rev. Microbiol. 2008;62:35–51. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

