Identification of extracellular miRNA in human cerebrospinal fluid by next-generation sequencing
- PMID: 23525801
- PMCID: PMC3677285
- DOI: 10.1261/rna.036863.112
Identification of extracellular miRNA in human cerebrospinal fluid by next-generation sequencing
Abstract
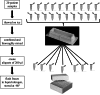
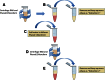
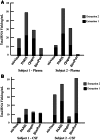
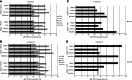
There has been a growing interest in using next-generation sequencing (NGS) to profile extracellular small RNAs from the blood and cerebrospinal fluid (CSF) of patients with neurological diseases, CNS tumors, or traumatic brain injury for biomarker discovery. Small sample volumes and samples with low RNA abundance create challenges for downstream small RNA sequencing assays. Plasma, serum, and CSF contain low amounts of total RNA, of which small RNAs make up a fraction. The purpose of this study was to maximize RNA isolation from RNA-limited samples and apply these methods to profile the miRNA in human CSF by small RNA deep sequencing. We systematically tested RNA isolation efficiency using ten commercially available kits and compared their performance on human plasma samples. We used RiboGreen to quantify total RNA yield and custom TaqMan assays to determine the efficiency of small RNA isolation for each of the kits. We significantly increased the recovery of small RNA by repeating the aqueous extraction during the phenol-chloroform purification in the top performing kits. We subsequently used the methods with the highest small RNA yield to purify RNA from CSF and serum samples from the same individual. We then prepared small RNA sequencing libraries using Illumina's TruSeq sample preparation kit and sequenced the samples on the HiSeq 2000. Not surprisingly, we found that the miRNA expression profile of CSF is substantially different from that of serum. To our knowledge, this is the first time that the small RNA fraction from CSF has been profiled using next-generation sequencing.
Figures


References
-
- Aldridge S, Hadfield J 2012. Introduction to miRNA profiling technologies and cross-platform comparison. Methods Mol Biol 822: 19–31 - PubMed
-
- Baraniskin A, Kuhnhenn J, Schlegel U, Chan A, Deckert M, Gold R, Maghnouj A, Zollner H, Reinacher-Schick A, Schmiegel W, et al. 2011. Identification of microRNAs in the cerebrospinal fluid as marker for primary diffuse large B-cell lymphoma of the central nervous system. Blood 117: 3140–3146 - PubMed
-
- Casalini P, Iorio MV 2010. MicroRNAs and future therapeutic applications in cancer. J BUON 14: S17–S22 - PubMed
-
- Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, et al. 2008. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 18: 997–1006 - PubMed
-
- Chen L, Yan HX, Yang W, Hu L, Yu LX, Liu Q, Li L, Huang DD, Ding J, Shen F, et al. 2009. The role of microRNA expression pattern in human intrahepatic cholangiocarcinoma. J Hepatol 50: 358–369 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
