Protein Structure Refinement through Structure Selection and Averaging from Molecular Dynamics Ensembles
- PMID: 23526422
- PMCID: PMC3603382
- DOI: 10.1021/ct300962x
Protein Structure Refinement through Structure Selection and Averaging from Molecular Dynamics Ensembles
Abstract
A molecular dynamics (MD) simulation based protocol for structure refinement of template-based model predictions is described. The protocol involves the application of restraints, ensemble averaging of selected subsets, interpolation between initial and refined structures, and assessment of refinement success. It is found that sub-microsecond MD-based sampling when combined with ensemble averaging can produce moderate but consistent refinement for most systems in the CASP targets considered here.
Keywords: force field; model selection; molecular dynamics simulation; scoring; structure prediction.
Figures

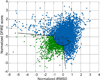
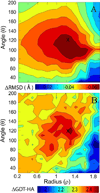
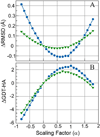
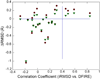
References
-
- Bradley P, Misura KMS, Baker D. Science. 2005;309:1868–1871. - PubMed
-
- Šali A, Blundell TL. J. Mol. Biol. 1993;234:779–815. - PubMed
-
- Lance BK, Deane CM, Wood GR. Bioinformatics. 2010;26:1849–1856. - PubMed
-
- Joo K, Lee J, Lee S, Seo J-H, Lee SJ, Lee J. Proteins: Struct., Funct., Bioinf. 2007;69:83–89. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
