Analysis of flavonoids and the flavonoid structural genes in brown fiber of upland cotton
- PMID: 23527031
- PMCID: PMC3602603
- DOI: 10.1371/journal.pone.0058820
Analysis of flavonoids and the flavonoid structural genes in brown fiber of upland cotton
Abstract
Background: As a result of changing consumer preferences, cotton (Gossypium Hirsutum L.) from varieties with naturally colored fibers is becoming increasingly sought after in the textile industry. The molecular mechanisms leading to colored fiber development are still largely unknown, although it is expected that the color is derived from flavanoids.
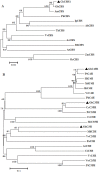
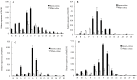
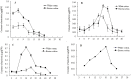
Experimental design: Firstly, four key genes of the flavonoid biosynthetic pathway in cotton (GhC4H, GhCHS, GhF3'H, and GhF3'5'H) were cloned and studied their expression profiles during the development of brown- and white cotton fibers by QRT-PCR. And then, the concentrations of four components of the flavonoid biosynthetic pathway, naringenin, quercetin, kaempferol and myricetin in brown- and white fibers were analyzed at different developmental stages by HPLC.
Result: The predicted proteins of the four flavonoid structural genes corresponding to these genes exhibit strong sequence similarity to their counterparts in various plant species. Transcript levels for all four genes were considerably higher in developing brown fibers than in white fibers from a near isogenic line (NIL). The contents of four flavonoids (naringenin, quercetin, kaempferol and myricetin) were significantly higher in brown than in white fibers and corresponding to the biosynthetic gene expression levels.
Conclusions: Flavonoid structural gene expression and flavonoid metabolism are important in the development of pigmentation in brown cotton fibers.
Conflict of interest statement
Figures



References
-
- Vreeland JJM (1999) The revival of colored cotton. Scientific American 280: 112–119.
-
- Qiu XM (2004) Research progress and prospects on naturally-colored cotton,Cotton Sciences. 16: 249–254.
-
- Zhao XQ, Wang XD (2005) Composition Analysis of Pigment in Colored Cotton Fiber. Acta Agronomica Sinice 4: 456–462.
-
- Kohel RJ (1985) Genetic analysis of fiber color variants in cotton. Crop Science 25: 793–797.
-
- Zhan SH, Li ZP, Lin Y, Cai YP (2008) Quantitative analysis on the inherited characteristics of naturally colored brown cotton fiber color. Chinese Agricultural Science Bulletin 24: 146–148.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

