Crystal structure of 70S ribosome with both cognate tRNAs in the E and P sites representing an authentic elongation complex
- PMID: 23527033
- PMCID: PMC3602588
- DOI: 10.1371/journal.pone.0058829
Crystal structure of 70S ribosome with both cognate tRNAs in the E and P sites representing an authentic elongation complex
Abstract
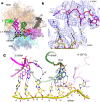
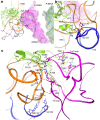
During the translation cycle, a cognate deacylated tRNA can only move together with the codon into the E site. We here present the first structure of a cognate tRNA bound to the ribosomal E site resulting from translocation by EF-G, in which an entire L1 stalk (L1 protein and L1 rRNA) interacts with E-site tRNA (E-tRNA), representing an authentic ribosome elongation complex. Our results revealed that the Watson-Crick base pairing is formed at the first and second codon-anticodon positions in the E site in the ribosome elongation complex, whereas the codon-anticodon interaction in the third position is indirect. Analysis of the observed conformations of mRNA and E-tRNA suggests that the ribosome intrinsically has the potential to form codon-anticodon interaction in the E site, independently of the mRNA configuration. We also present a detailed description of the biologically relevant position of the entire L1 stalk and its interacting cognate E-tRNA, which provides a better understanding of the structural basis for translation elongation. Furthermore, to gain insight into translocation, we report the positioning of protein L6 contacting EF-G, as well as the conformational change of the C-terminal tail of protein S13 in the decoding center.
Conflict of interest statement
Figures



References
-
- Schmeing TM, Ramakrishnan V (2009) What recent ribosome structures have revealed about the mechanism of translation. Nature 461: 1234–1242. - PubMed
-
- Frank J, Agrawal RK (2000) A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 406: 319–322. - PubMed
-
- Moazed D, Noller HF (1989) Intermediate states in the movement of transfer RNA in the ribosome. Nature 342: 142–148. - PubMed
-
- Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W (1997) Hydrolysis of GTP by elongation factor G drives tRNA movement on the ribosome. Nature 385: 37–41. - PubMed
-
- Bodley JW, Zieve FJ, Lin L, Zieve ST (1970) Studies on translocation. 3. Conditions necessary for the formation and detection of a stable ribosome-G factor-guanosine diphosphate complex in the presence of fusidic acid. J Biol Chem 245: 5656–5661. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

