DNA polymerase ε and δ exonuclease domain mutations in endometrial cancer
- PMID: 23528559
- PMCID: PMC3690967
- DOI: 10.1093/hmg/ddt131
DNA polymerase ε and δ exonuclease domain mutations in endometrial cancer
Abstract
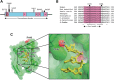
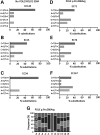
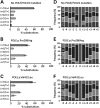
Accurate duplication of DNA prior to cell division is essential to suppress mutagenesis and tumour development. The high fidelity of eukaryotic DNA replication is due to a combination of accurate incorporation of nucleotides into the nascent DNA strand by DNA polymerases, the recognition and removal of mispaired nucleotides (proofreading) by the exonuclease activity of DNA polymerases δ and ε, and post-replication surveillance and repair of newly synthesized DNA by the mismatch repair (MMR) apparatus. While the contribution of defective MMR to neoplasia is well recognized, evidence that faulty DNA polymerase activity is important in cancer development has been limited. We have recently shown that germline POLE and POLD1 exonuclease domain mutations (EDMs) predispose to colorectal cancer (CRC) and, in the latter case, to endometrial cancer (EC). Somatic POLE mutations also occur in 5-10% of sporadic CRCs and underlie a hypermutator, microsatellite-stable molecular phenotype. We hypothesized that sporadic ECs might also acquire somatic POLE and/or POLD1 mutations. Here, we have found that missense POLE EDMs with good evidence of pathogenic effects are present in 7% of a set of 173 endometrial cancers, although POLD1 EDMs are uncommon. The POLE mutations localized to highly conserved residues and were strongly predicted to affect proofreading. Consistent with this, POLE-mutant tumours were hypermutated, with a high frequency of base substitutions, and an especially large relative excess of G:C>T:A transversions. All POLE EDM tumours were microsatellite stable, suggesting that defects in either DNA proofreading or MMR provide alternative mechanisms to achieve genomic instability and tumourigenesis.
Figures
References
-
- Kunkel T.A. DNA replication fidelity. J. Biol. Chem. 2004;279:16895–16898. doi:10.1074/jbc.R400006200. - DOI - PubMed
-
- Pursell Z.F., Isoz I., Lundström E-B., Johansson E., Kunkel T.A. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–130. doi:10.1126/science.1144067. - DOI - PMC - PubMed
-
- McElhinny S.A., Gordenin D.A., Stith C.M., Burgers P.M.J., Kunkel T.A. Division of labor at the eukaryotic replication fork. Mol. Cell. 2008;30:137–144. doi:10.1016/j.molcel.2008.02.022. - DOI - PMC - PubMed
-
- Larrea A.A., Lujan S.A., Nick McElhinny S.A., Mieczkowski P.A., Resnick M.A., Gordenin D.A., Kunkel T.A. Genome-wide model for the normal eukaryotic DNA replication fork. Proc. Natl Acad. Sci. USA. 2010;107:17674–17679. doi:10.1073/pnas.1010178107. - DOI - PMC - PubMed
-
- Miyabe I., Kunkel T.A., Carr A.M. The major roles of DNA polymerases epsilon and delta at the eukaryotic replication fork are evolutionarily conserved. PLoS Genet. 2011;7:e1002407. doi:10.1371/journal.pgen.1002407. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

