Role and regulation of PDGFRα signaling in liver development and regeneration
- PMID: 23529017
- PMCID: PMC3644730
- DOI: 10.1016/j.ajpath.2013.01.047
Role and regulation of PDGFRα signaling in liver development and regeneration
Abstract
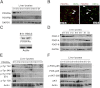
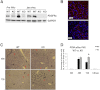
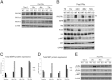
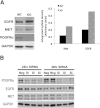
Aberrant platelet-derived growth factor receptor-α (PDGFRα) signaling is evident in a subset of hepatocellular cancers (HCCs). However, its role and regulation in hepatic physiology remains elusive. In the current study, we examined PDGFRα signaling in liver development and regeneration. We identified notable PDGFRα activation in hepatic morphogenesis that, when interrupted by PDGFRα-blocking antibody, led to decreased hepatoblast proliferation and survival in embryonic liver cultures. We also identified temporal PDGFRα overexpression, which is regulated by epidermal growth factor (EGF) and tumor necrosis factor α, and its activation at 3 to 24 hours after partial hepatectomy. Through generation of hepatocyte-specific PDGFRA knockout (KO) mice that lack an overt phenotype, we show that absent PDGFRα compromises extracelluar signal-regulated kinases and AKT activation 3 hours after partial hepatectomy, which, however, is alleviated by temporal compensatory increases in the EGF receptor (EGFR) and the hepatocyte growth factor receptor (Met) expression and activation along with rebound activation of extracellular signal-regulated kinases and AKT at 24 hours. These untimely increases in EGFR and Met allow for normal hepatocyte proliferation at 48 hours in KO, which, however, are aberrantly prolonged up to 72 hours. Intriguingly, such compensation also was visible in primary KO hepatocyte cultures but not in HCC cells after siRNA-mediated PDGFRα knockdown. Thus, temporal activation of PDGFRα in liver development is important in hepatic morphogenesis. In liver regeneration, despite increased signaling, PDGFRα is dispensable owing to EGFR and Met compensation, which is unique to normal hepatocytes but not HCC cells.
Copyright © 2013 American Society for Investigative Pathology. Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Claesson-Welsh L. Platelet-derived growth factor receptor signals. J Biol Chem. 1994;269:32023–32026. - PubMed
-
- Hoch R.V., Soriano P. Roles of PDGF in animal development. Development. 2003;130:4769–4784. - PubMed
-
- Heldin C.H., Ostman A., Ronnstrand L. Signal transduction via platelet-derived growth factor receptors. Biochim Biophys Acta. 1998;1378:F79–F113. - PubMed
-
- Franke T.F., Yang S.I., Chan T.O., Datta K., Kazlauskas A., Morrison D.K., Kaplan D.R., Tsichlis P.N. The protein kinase encoded by the Akt proto-oncogene is a target of the PDGF-activated phosphatidylinositol 3-kinase. Cell. 1995;81:727–736. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

