Impact of primary elvitegravir resistance-associated mutations in HIV-1 integrase on drug susceptibility and viral replication fitness
- PMID: 23529738
- PMCID: PMC3716146
- DOI: 10.1128/AAC.02568-12
Impact of primary elvitegravir resistance-associated mutations in HIV-1 integrase on drug susceptibility and viral replication fitness
Abstract
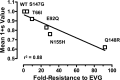
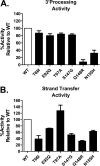
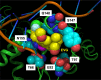
Elvitegravir (EVG) is an effective HIV-1 integrase (IN) strand transfer inhibitor (INSTI) in advanced clinical development. Primary INSTI resistance-associated mutations (RAMs) at six IN positions have been identified in HIV-1-infected patients failing EVG-containing regimens in clinical studies: T66I/A/K, E92Q/G, T97A, S147G, Q148R/H/K, and N155H. In this study, the effect of these primary IN mutations, alone and in combination, on susceptibility to the INSTIs EVG, raltegravir (RAL), and dolutegravir (DTG); IN enzyme activities; and viral replication fitness was characterized. Recombinant viruses containing the six most common mutations exhibited a range of reduced EVG susceptibility: 92-fold for Q148R, 30-fold for N155H, 26-fold for E92Q, 10-fold for T66I, 4-fold for S147G, and 2-fold for T97A. Less commonly observed primary IN mutations also showed a range of reduced EVG susceptibilities: 40- to 94-fold for T66K and Q148K and 5- to 10-fold for T66A, E92G, and Q148H. Some primary IN mutations exhibited broad cross-resistance between EVG and RAL (T66K, E92Q, Q148R/H/K, and N155H), while others retained susceptibility to RAL (T66I/A, E92G, T97A, and S147G). Dual combinations of primary IN mutations further reduced INSTI susceptibility, replication capacity, and viral fitness relative to either mutation alone. Susceptibility to DTG was retained by single primary IN mutations but reduced by dual mutation combinations with Q148R. Primary EVG RAMs also diminished IN enzymatic activities, concordant with their structural proximity to the active site. Greater reductions in viral fitness of dual mutation combinations may explain why some primary INSTI RAMs do not readily coexist on the same HIV-1 genome but rather establish independent pathways of resistance to EVG.
Figures

References
-
- Thompson MA, Aberg JA, Hoy JF, Telenti A, Benson C, Cahn P, Eron JJ, Gunthard HF, Hammer SM, Reiss P, Richman DD, Rizzardini G, Thomas DL, Jacobsen DM, Volberding PA. 2012. Antiretroviral treatment of adult HIV infection: 2012 recommendations of the International Antiviral Society-USA Panel. JAMA 308:387–402 - PubMed
-
- Engelman A, Mizuuchi K, Craigie R. 1991. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell 67:1211–1221 - PubMed
-
- Eron JJ, Jr, Rockstroh JK, Reynes J, Andrade-Villanueva J, Ramalho-Madruga JV, Bekker LG, Young B, Katlama C, Gatell-Artigas JM, Arribas JR, Nelson M, Campbell H, Zhao J, Rodgers AJ, Rizk ML, Wenning L, Miller MD, Hazuda D, DiNubile MJ, Leavitt R, Isaacs R, Robertson MN, Sklar P, Nguyen BY. 2011. Raltegravir once daily or twice daily in previously untreated patients with HIV-1: a randomised, active-controlled, phase 3 non-inferiority trial. Lancet Infect. Dis. 11:907–915 - PubMed
-
- Steigbigel RT, Cooper DA, Teppler H, Eron JJ, Gatell JM, Kumar PN, Rockstroh JK, Schechter M, Katlama C, Markowitz M, Yeni P, Loutfy MR, Lazzarin A, Lennox JL, Clotet B, Zhao J, Wan H, Rhodes RR, Strohmaier KM, Barnard RJ, Isaacs RD, Nguyen BY. 2010. Long-term efficacy and safety of raltegravir combined with optimized background therapy in treatment-experienced patients with drug-resistant HIV infection: week 96 results of the BENCHMRK 1 and 2 phase III trials. Clin. Infect. Dis. 50:605–612 - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

