The hyaluronan receptor for endocytosis (HARE) activates NF-κB-mediated gene expression in response to 40-400-kDa, but not smaller or larger, hyaluronans
- PMID: 23530033
- PMCID: PMC3656264
- DOI: 10.1074/jbc.M112.442889
The hyaluronan receptor for endocytosis (HARE) activates NF-κB-mediated gene expression in response to 40-400-kDa, but not smaller or larger, hyaluronans
Abstract
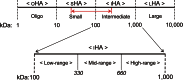
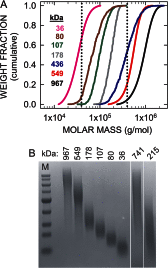
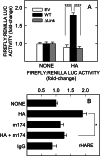
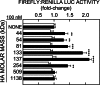
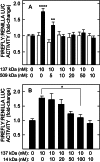
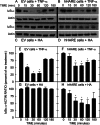
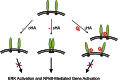
The hyaluronan (HA) receptor for endocytosis (HARE; Stabilin-2) binds and clears 14 different ligands, including HA and heparin, via clathrin-mediated endocytosis. HA binding to HARE stimulates ERK1/2 activation (Kyosseva, S. V., Harris, E. N., and Weigel, P. H. (2008) J. Biol. Chem. 283, 15047-15055). To assess a possible HA size dependence for signaling, we tested purified HA fractions of different weight-average molar mass and with narrow size distributions and Select-HA(TM) for stimulation of HARE-mediated gene expression using an NF-κB promoter-driven luciferase reporter system. Human HARE-mediated gene expression was stimulated in a dose-dependent manner with small HA (sHA) >40 kDa and intermediate HA (iHA) <400 kDa. The hyperbolic dose response saturated at 20-50 nM with an apparent K(m) ~10 nM, identical to the Kd for HA-HARE binding. Activation was not detected with oligomeric HA (oHA), sHA <40 kDa, iHA >400 kDa, or large HA (lHA). Similar responses occurred with rat HARE. Activation by sHA-iHA was blocked by excess nonsignaling sHA, iHA, or lHA, deletion of the HA-binding LINK domain, or HA-blocking antibody. Endogenous NF-κB activation also occurred in the absence of luciferase plasmids, as assessed by degradation of IκB-α. ERK1/2 activation was also HA size-dependent. The results show that HA-HARE interactions stimulate NF-κB-activated gene expression and that HARE senses a narrow size range of HA degradation products. We propose a model in which optimal length HA binds multiple HARE proteins to allow cytoplasmic domain interactions that stimulate intracellular signaling. This HARE signaling system during continuous HA clearance could monitor the homeostasis of tissue biomatrix turnover throughout the body.
Keywords: Cell Signaling; ERK1/2; Glycosaminoglycan; Hyaluronate; Scavenger Receptor; Select-HA; Signal Transduction; Stabilin-2; Stress Response; Transcription.
Figures





References
-
- Laurent T. C., Laurent U. B., Fraser J. R. (1996) The structure and function of hyaluronan: An overview. Immunol. Cell Biol. 74, A1–A7 - PubMed
-
- Toole B. P. (1990) Hyaluronan and its binding proteins, the hyaladherins. Curr. Opin. Cell Biol. 2, 839–844 - PubMed
-
- Day A. J., Prestwich G. D. (2002) Hyaluronan-binding proteins: tying up the giant. J. Biol. Chem. 277, 4585–4588 - PubMed
-
- Laurent T. C., Fraser J. R. (1992) Hyaluronan. FASEB J. 6, 2397–2404 - PubMed
-
- Knudson C. B., Knudson W. (1993) Hyaluronan-binding proteins in development, tissue homeostasis, and disease. FASEB J. 7, 1233–1241 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

