Distinctive contributions of the ribosomal P-site elements m2G966, m5C967 and the C-terminal tail of the S9 protein in the fidelity of initiation of translation in Escherichia coli
- PMID: 23530111
- PMCID: PMC3643588
- DOI: 10.1093/nar/gkt175
Distinctive contributions of the ribosomal P-site elements m2G966, m5C967 and the C-terminal tail of the S9 protein in the fidelity of initiation of translation in Escherichia coli
Abstract
The accuracy of pairing of the anticodon of the initiator tRNA (tRNA(fMet)) and the initiation codon of an mRNA, in the ribosomal P-site, is crucial for determining the translational reading frame. However, a direct role of any ribosomal element(s) in scrutinizing this pairing is unknown. The P-site elements, m(2)G966 (methylated by RsmD), m(5)C967 (methylated by RsmB) and the C-terminal tail of the protein S9 lie in the vicinity of tRNA(fMet). We investigated the role of these elements in initiation from various codons, namely, AUG, GUG, UUG, CUG, AUA, AUU, AUC and ACG with tRNA(fMet(CAU) (tRNA(fMet) with CAU anticodon); CAC and CAU with tRNA(fMet(GUG); UAG with tRNA(fMet(CAU) ; UAC with tRNA(fMet(GUG) ; and AUC with tRNA(fMet(GUG) using in vivo and computational methods. Although RsmB deficiency did not impact initiation from most codons, RsmD deficiency increased initiation from AUA, CAC and CAU (2- to 3.6-fold). Deletion of the S9 C-terminal tail resulted in poorer initiation from UUG, GUG and CUG, but in increased initiation from CAC, CAU and UAC codons (up to 4-fold). Also, the S9 tail suppressed initiation with tRNA(fMet(CAU) lacking the 3GC base pairs in the anticodon stem. These observations suggest distinctive roles of 966/967 methylations and the S9 tail in initiation.
Figures
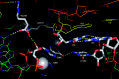
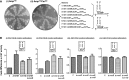
 (encoded by metYGUG) as assessed by growth on Cm plate (A). Saturated cultures of E. coli SA and its ΔrsmB, ΔrsmD, and ΔrsmB ΔrsmD derivatives harbouring pCATCAUmetYGUG (sectors 1–4) or pCATCACmetYGUG (sectors 5–8) were streaked on Amp100 (left) or Amp100 Cm250 (right) and incubated at 37°C for ∼15 h. CAT assays (B) to assess initiation activities of CAC:GUG (panel i), CAU:GUG (panel ii), AUG:CAU (panel iii) and UAG:CUA (panel iv) codon:anticodon pairs. Star symbols in the boxes indicate G966 (anticodon proximal) and C967. Fold differences in the CAT activities with respect to SA (C, taken as 1) in its ΔrsmB, ΔrsmD, and ΔrsmB ΔrsmD derivatives are shown. For reference, the average CAT activities in SA (C) for the CAC:GUG, CAU:GUG, AUG:CAU and UAG:CUA codon:anticodon pairs were 1462 ± 374, 493 ± 64, 7314 ± 923 and 5246 ± 763 pmol Cm acetylated per 1 μg of total cell-free extract in 20 min at 37°C, respectively.
(encoded by metYGUG) as assessed by growth on Cm plate (A). Saturated cultures of E. coli SA and its ΔrsmB, ΔrsmD, and ΔrsmB ΔrsmD derivatives harbouring pCATCAUmetYGUG (sectors 1–4) or pCATCACmetYGUG (sectors 5–8) were streaked on Amp100 (left) or Amp100 Cm250 (right) and incubated at 37°C for ∼15 h. CAT assays (B) to assess initiation activities of CAC:GUG (panel i), CAU:GUG (panel ii), AUG:CAU (panel iii) and UAG:CUA (panel iv) codon:anticodon pairs. Star symbols in the boxes indicate G966 (anticodon proximal) and C967. Fold differences in the CAT activities with respect to SA (C, taken as 1) in its ΔrsmB, ΔrsmD, and ΔrsmB ΔrsmD derivatives are shown. For reference, the average CAT activities in SA (C) for the CAC:GUG, CAU:GUG, AUG:CAU and UAG:CUA codon:anticodon pairs were 1462 ± 374, 493 ± 64, 7314 ± 923 and 5246 ± 763 pmol Cm acetylated per 1 μg of total cell-free extract in 20 min at 37°C, respectively.
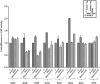
 from various codons. Star symbols in the box indicate G966 (anticodon proximal) and C967. Fold differences in the CAT activities with respect to SA (C, taken as 1) in its ΔrsmB, ΔrsmD, and ΔrsmB ΔrsmD derivatives are shown. For reference, the average CAT activities in the SA control (C) for the AUG, GUG, UUG, CUG, AUA, AUU, AUC and ACG constructs were 7314 ± 923, 3629 ± 492, 4019 ± 1000, 906 ± 132, 238 ± 33, 301 ± 32, 352.5 ± 38 and 48 ± 15 pmol Cm acetylated per 1 μg of total cell-free extract in 20 min at 37°C, respectively.
from various codons. Star symbols in the box indicate G966 (anticodon proximal) and C967. Fold differences in the CAT activities with respect to SA (C, taken as 1) in its ΔrsmB, ΔrsmD, and ΔrsmB ΔrsmD derivatives are shown. For reference, the average CAT activities in the SA control (C) for the AUG, GUG, UUG, CUG, AUA, AUU, AUC and ACG constructs were 7314 ± 923, 3629 ± 492, 4019 ± 1000, 906 ± 132, 238 ± 33, 301 ± 32, 352.5 ± 38 and 48 ± 15 pmol Cm acetylated per 1 μg of total cell-free extract in 20 min at 37°C, respectively.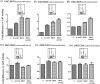
 ). A tail ending with SKR in the box on the top right indicates ribosomal protein S9. Fold differences in the CAT activities with respect to SA (C, taken as 1) in its ΔrsmD, S9Δ3, and S9Δ3 ΔrsmD derivatives are shown. For reference, the average CAT activities in SA strain (C, taken as 1) for the AUG, GUG, UUG, CUG, AUA, AUU, AUC and ACG constructs were 7838 ± 610, 3198 ± 378, 3788 ± 334, 327 ± 15, 313 ± 19, 346 ± 34, 631 ± 71.6 and 78 ± 9 pmol Cm acetylated per 1 μg of total cell-free extract in 20 min at 37°C, respectively.
). A tail ending with SKR in the box on the top right indicates ribosomal protein S9. Fold differences in the CAT activities with respect to SA (C, taken as 1) in its ΔrsmD, S9Δ3, and S9Δ3 ΔrsmD derivatives are shown. For reference, the average CAT activities in SA strain (C, taken as 1) for the AUG, GUG, UUG, CUG, AUA, AUU, AUC and ACG constructs were 7838 ± 610, 3198 ± 378, 3788 ± 334, 327 ± 15, 313 ± 19, 346 ± 34, 631 ± 71.6 and 78 ± 9 pmol Cm acetylated per 1 μg of total cell-free extract in 20 min at 37°C, respectively.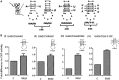
 . Various mutations in the anticodon stem are as shown (A). Fold differences in the CAT activities with respect to SA (C, taken as 1) in its S9Δ3 derivatives are shown (B) for 1GC (AU, panel i), 1GC (GU, panel ii), 2GC (AU/GU, panel iii) and the 3GC mutant (panel iv). For reference, the average CAT activities in SA for the UAG:CUA/AU, UAG:CUA/GU, UAG:CUA/AUGU and UAG:CUA/3GC constructs were 2970 ± 900, 888 ± 145, 32 ± 6, 238 ± 33 and 88 ± 19 pmol Cm acetylated per 1 μg of total cell-free extract in 20 min at 37°C, respectively.
. Various mutations in the anticodon stem are as shown (A). Fold differences in the CAT activities with respect to SA (C, taken as 1) in its S9Δ3 derivatives are shown (B) for 1GC (AU, panel i), 1GC (GU, panel ii), 2GC (AU/GU, panel iii) and the 3GC mutant (panel iv). For reference, the average CAT activities in SA for the UAG:CUA/AU, UAG:CUA/GU, UAG:CUA/AUGU and UAG:CUA/3GC constructs were 2970 ± 900, 888 ± 145, 32 ± 6, 238 ± 33 and 88 ± 19 pmol Cm acetylated per 1 μg of total cell-free extract in 20 min at 37°C, respectively.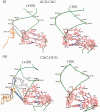

References
-
- Ogle JM, Brodersen DE, Clemons WM, Jr, Tarry MJ, Carter AP, Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. - PubMed
-
- Sacerdot C, Chiaruttini C, Engst K, Graffe M, Milet M, Mathy N, Dondon J, Springer M. The role of the AUU initiation codon in the negative feedback regulation of the gene for translation initiation factor IF3 in Escherichia coli. Mol. Microbiol. 1996;21:331–346. - PubMed
-
- Selmer M, Dunham CM, Murphy FV, IV, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

