De novo centromere formation on a chromosome fragment in maize
- PMID: 23530217
- PMCID: PMC3625319
- DOI: 10.1073/pnas.1303944110
De novo centromere formation on a chromosome fragment in maize
Abstract
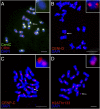
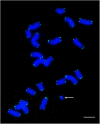
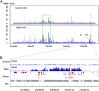
The centromere is the part of the chromosome that organizes the kinetochore, which mediates chromosome movement during mitosis and meiosis. A small fragment from chromosome 3, named Duplication 3a (Dp3a), was described from UV-irradiated materials by Stadler and Roman in the 1940s [Stadler LJ, Roman H (1948) Genetics 33(3):273-303]. The genetic behavior of Dp3a is reminiscent of a ring chromosome, but fluoresecent in situ hybridization detected telomeres at both ends, suggesting a linear structure. This small chromosome has no detectable canonical centromeric sequences, but contains a site with protein features of functional centromeres such as CENH3, the centromere specific H3 histone variant, and CENP-C, a foundational kinetochore protein, suggesting the de novo formation of a centromere on the chromatin fragment. To examine the sequences associated with CENH3, chromatin immunoprecipitation was carried out with anti-CENH3 antibodies using material from young seedlings with and without the Dp3a chromosome. A novel peak was detected from the ChIP-Sequencing reads of the Dp3a sample. The peak spanned 350 kb within the long arm of chromosome 3 covering 22 genes. Collectively, these results define the behavior and molecular features of de novo centromere formation in the Dp3a chromosome, which may shed light on the initiation of new centromere sites during evolution.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Sequential de novo centromere formation and inactivation on a chromosomal fragment in maize.Proc Natl Acad Sci U S A. 2015 Mar 17;112(11):E1263-71. doi: 10.1073/pnas.1418248112. Epub 2015 Mar 2. Proc Natl Acad Sci U S A. 2015. PMID: 25733907 Free PMC article.
-
Recurrent establishment of de novo centromeres in the pericentromeric region of maize chromosome 3.Chromosome Res. 2017 Oct;25(3-4):299-311. doi: 10.1007/s10577-017-9564-x. Epub 2017 Aug 22. Chromosome Res. 2017. PMID: 28831743
-
Sequences associated with A chromosome centromeres are present throughout the maize B chromosome.Chromosoma. 2005 Feb;113(7):337-49. doi: 10.1007/s00412-004-0319-z. Epub 2004 Dec 7. Chromosoma. 2005. PMID: 15586285
-
Dicentric chromosome formation and epigenetics of centromere formation in plants.J Genet Genomics. 2012 Mar 20;39(3):125-30. doi: 10.1016/j.jgg.2012.01.006. Epub 2012 Feb 14. J Genet Genomics. 2012. PMID: 22464471 Review.
-
Genetic and epigenetic effects on centromere establishment.Chromosoma. 2020 Mar;129(1):1-24. doi: 10.1007/s00412-019-00727-3. Epub 2019 Nov 28. Chromosoma. 2020. PMID: 31781852 Review.
Cited by
-
Rapid Birth or Death of Centromeres on Fragmented Chromosomes in Maize.Plant Cell. 2020 Oct;32(10):3113-3123. doi: 10.1105/tpc.20.00389. Epub 2020 Aug 18. Plant Cell. 2020. PMID: 32817254 Free PMC article.
-
Meiotic behavior of small chromosomes in maize.Front Plant Sci. 2013 Dec 17;4:505. doi: 10.3389/fpls.2013.00505. Front Plant Sci. 2013. PMID: 24381575 Free PMC article. Review.
-
Meiotic Studies on Combinations of Chromosomes With Different Sized Centromeres in Maize.Front Plant Sci. 2018 Jun 13;9:785. doi: 10.3389/fpls.2018.00785. eCollection 2018. Front Plant Sci. 2018. PMID: 29951076 Free PMC article.
-
Dynamic epigenetic states of maize centromeres.Front Plant Sci. 2015 Oct 26;6:904. doi: 10.3389/fpls.2015.00904. eCollection 2015. Front Plant Sci. 2015. PMID: 26579154 Free PMC article. Review.
-
De Novo Centromere Formation and Centromeric Sequence Expansion in Wheat and its Wide Hybrids.PLoS Genet. 2016 Apr 25;12(4):e1005997. doi: 10.1371/journal.pgen.1005997. eCollection 2016 Apr. PLoS Genet. 2016. PMID: 27110907 Free PMC article.
References
-
- Flemming W. Zellsubstanz, Kern und Zelltheilung [Cell substance, nucleus and cell division] Leipzig, Germany: FCW Vogel; 1882. German.
-
- Henikoff S, Ahmad K, Malik HS. The centromere paradox: Stable inheritance with rapidly evolving DNA. Science. 2001;293(5532):1098–1102. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

