A systematic comparison of the MetaCyc and KEGG pathway databases
- PMID: 23530693
- PMCID: PMC3665663
- DOI: 10.1186/1471-2105-14-112
A systematic comparison of the MetaCyc and KEGG pathway databases
Abstract
Background: The MetaCyc and KEGG projects have developed large metabolic pathway databases that are used for a variety of applications including genome analysis and metabolic engineering. We present a comparison of the compound, reaction, and pathway content of MetaCyc version 16.0 and a KEGG version downloaded on Feb-27-2012 to increase understanding of their relative sizes, their degree of overlap, and their scope. To assess their overlap, we must know the correspondences between compounds, reactions, and pathways in MetaCyc, and those in KEGG. We devoted significant effort to computational and manual matching of these entities, and we evaluated the accuracy of the correspondences.
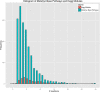
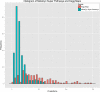
Results: KEGG contains 179 module pathways versus 1,846 base pathways in MetaCyc; KEGG contains 237 map pathways versus 296 super pathways in MetaCyc. KEGG pathways contain 3.3 times as many reactions on average as do MetaCyc pathways, and the databases employ different conceptualizations of metabolic pathways. KEGG contains 8,692 reactions versus 10,262 for MetaCyc. 6,174 KEGG reactions are components of KEGG pathways versus 6,348 for MetaCyc. KEGG contains 16,586 compounds versus 11,991 for MetaCyc. 6,912 KEGG compounds act as substrates in KEGG reactions versus 8,891 for MetaCyc. MetaCyc contains a broader set of database attributes than does KEGG, such as relationships from a compound to enzymes that it regulates, identification of spontaneous reactions, and the expected taxonomic range of metabolic pathways. MetaCyc contains many pathways not found in KEGG, from plants, fungi, metazoa, and actinobacteria; KEGG contains pathways not found in MetaCyc, for xenobiotic degradation, glycan metabolism, and metabolism of terpenoids and polyketides. MetaCyc contains fewer unbalanced reactions, which facilitates metabolic modeling such as using flux-balance analysis. MetaCyc includes generic reactions that may be instantiated computationally.
Conclusions: KEGG contains significantly more compounds than does MetaCyc, whereas MetaCyc contains significantly more reactions and pathways than does KEGG, in particular KEGG modules are quite incomplete. The number of reactions occurring in pathways in the two DBs are quite similar.
Figures
References
-
- Caspi R, Altman T, Dale JM, Dreher K, Fulcher CA, Gilham F, Kaipa P, Karthikeyan AS, Kothari A, Krummenacker M, Latendresse M, Mueller LA, Paley S, Popescu L, Pujar A, Shearer AG, Zhang P, Karp PD. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nuc Acids Res. 2010;38:D473–D479. doi: 10.1093/nar/gkp875. advanced access [ http://dx.doi.org/10.1093/nar/gkp875] - DOI - PMC - PubMed
-
- Caspi R, Altman T, Dreher K, Fulcher CA, Subhraveti P, Keseler I, Kothari A, Krummenacker M, Latendresse M, Mueller LA, Ong Q, Paley S, Pujar A, Shearer AG, Travers M, Weerasinghe D, Zhang P, Karp PD. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nuc Acids Res. 2012;40:D742–D753. doi: 10.1093/nar/gkr1014. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

