Evolution of cryptic gene pools in Hypericum perforatum: the influence of reproductive system and gene flow
- PMID: 23532046
- PMCID: PMC3662511
- DOI: 10.1093/aob/mct065
Evolution of cryptic gene pools in Hypericum perforatum: the influence of reproductive system and gene flow
Abstract
Background and aims: Hypericum perforatum (St. John's wort) is a widespread Eurasian perennial plant species with remarkable variation in its morphology, ploidy and breeding system, which ranges from sex to apomixis. Here, hypotheses on the evolutionary origin of St. John's wort are tested and contrasted with the subsequent history of interspecific gene flow.
Methods: Extensive field collections were analysed for quantitative morphological variation, ploidy, chromosome numbers and genetic diversity using nuclear (amplified fragment length polymorphism) and plastid (trnL-trnF) markers. The mode of reproduction was analysed by FCSS (flow cytometric seed screen).
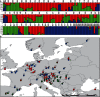
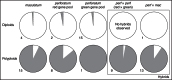
Key results: It is demonstrated that H. perforatum is not of hybrid origin, and for the first time wild diploid populations are documented. Pseudogamous facultative apomictic reproduction is prevalent in the polyploids, whereas diploids are predominantly sexual, a phenomenon which also characterizes its sister species H. maculatum. Both molecular markers characterize identical major gene pools, distinguishing H. perforatum from H. maculatum and two genetic groups in H. perforatum. All three gene pools are in close geographical contact. Extensive gene flow and hybridization throughout Europe within and between gene pools and species is exemplified by the molecular data and confirmed by morphometric analyses.
Conclusions: Hypericum perforatum is of a single evolutionary origin and later split into two major gene pools. Subsequently, independent and recurrent polyploidization occurred in all lineages and was accompanied by substantial gene flow within and between H. perforatum and H. maculatum. These processes are highly influenced by the reproductive system in both species, with a switch to predominantly apomictic reproduction in polyploids, irrespective of their origin.
Keywords: Apomixis; Hypericum maculatum; Hypericum perforatum; St. John's wort; evolution; hybridization; reproductive mode.
Figures



Similar articles
-
De novo sequencing of the Hypericum perforatum L. flower transcriptome to identify potential genes that are related to plant reproduction sensu lato.BMC Genomics. 2015 Mar 31;16(1):254. doi: 10.1186/s12864-015-1439-y. BMC Genomics. 2015. PMID: 25887758 Free PMC article.
-
Biogeographic variation in genetic variability, apomixis expression and ploidy of St. John's wort (Hypericum perforatum) across its native and introduced range.Ann Bot. 2014 Feb;113(3):417-27. doi: 10.1093/aob/mct268. Epub 2013 Dec 15. Ann Bot. 2014. PMID: 24344138 Free PMC article.
-
Cryptic gene pools in the Hypericum perforatum-H. maculatum complex: diploid persistence versus trapped polyploid melting.Ann Bot. 2017 Nov 28;120(6):955-966. doi: 10.1093/aob/mcx110. Ann Bot. 2017. PMID: 29182722 Free PMC article.
-
St John's wort (Hypericum perforatum) in major depression.J Clin Psychiatry. 2009;70 Suppl 5:23-7. doi: 10.4088/JCP.8157su1c.05. J Clin Psychiatry. 2009. PMID: 19909690 Review.
-
The production of hypericins and hyperforin by in vitro cultures of St. John's wort (Hypericum perforatum).Biotechnol Appl Biochem. 2004 Feb;39(Pt 1):71-81. doi: 10.1042/BA20030144. Biotechnol Appl Biochem. 2004. PMID: 14521510 Review.
Cited by
-
Separation in flowering time contributes to the maintenance of sympatric cryptic plant lineages.Ecol Evol. 2015 Jun;5(11):2172-84. doi: 10.1002/ece3.1481. Epub 2015 May 8. Ecol Evol. 2015. PMID: 26078854 Free PMC article.
-
De novo sequencing of the Hypericum perforatum L. flower transcriptome to identify potential genes that are related to plant reproduction sensu lato.BMC Genomics. 2015 Mar 31;16(1):254. doi: 10.1186/s12864-015-1439-y. BMC Genomics. 2015. PMID: 25887758 Free PMC article.
-
Geographical Parthenogenesis in Alpine and Arctic Plants.Plants (Basel). 2023 Feb 13;12(4):844. doi: 10.3390/plants12040844. Plants (Basel). 2023. PMID: 36840192 Free PMC article. Review.
-
Species delimitation and recognition in the Pediomelummegalanthum complex (Fabaceae) via multivariate morphometrics.PhytoKeys. 2015 Jan 13;(44):65-87. doi: 10.3897/phytokeys.44.8750. eCollection 2015. PhytoKeys. 2015. PMID: 25698894 Free PMC article.
-
Underexplored biodiversity of Eastern Mediterranean biota: systematics and evolutionary history of the genus Aubrieta (Brassicaceae).Ann Bot. 2017 Jan;119(1):39-57. doi: 10.1093/aob/mcw204. Epub 2016 Dec 10. Ann Bot. 2017. PMID: 27941091 Free PMC article.
References
-
- APG III. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Botanical Journal of the Linnean Society. 2009;161:105–121.
-
- Barcaccia G, Arzenton F, Sharbel TF, Varotto S, Parrini P, Lucchin M. Genetic diversity and reproductive biology in local populations of the facultative apomict Hypericum perforatum L. Heredity. 2006;96:322–334. - PubMed
-
- Bennett MD, Leitch IJ. Angiosperm DNA C-values database. 2010 (release 7·0, December 2010). http://www.kew.org/cvalues/
-
- Bonin A, Bellemain E, Eidesen P, Pompanon F, Brochmann C, Taberlet P. How to track and assess genotyping errors in population genetics studies. Molecular Ecology. 2004;13:3261–3273. - PubMed
-
- Brutovská R, Cellárová E, Schubert I. Cytogenetic characterization of three Hypericum species by in situ hybridization. Theoretical and Applied Genetics. 2000;101:46–50.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

