BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis
- PMID: 23532072
- PMCID: PMC3634682
- DOI: 10.1105/tpc.112.107904
BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis
Abstract
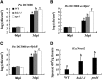
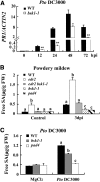
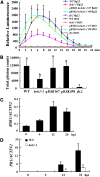
Pathogen-associated molecular pattern (PAMP)-trigged immunity (PTI) is the first defensive line of plant innate immunity and is mediated by pattern recognition receptors. Here, we show that a mutation in BR-SIGNALING KINASE1 (BSK1), a substrate of the brassinosteroid (BR) receptor BRASSINOSTEROID INSENSITIVE1, suppressed the powdery mildew resistance caused by a mutation in ENHANCED DISEASE RESISTANCE2, which negatively regulates powdery mildew resistance and programmed cell death, in Arabidopsis thaliana. A loss-of-function bsk1 mutant displayed enhanced susceptibility to virulent and avirulent pathogens, including Golovinomyces cichoracearum, Pseudomonas syringae, and Hyaloperonospora arabidopsidis. The bsk1 mutant also accumulated lower levels of salicylic acid upon infection with G. cichoracearum and P. syringae. BSK1 belongs to a receptor-like cytoplasmic kinase family and displays kinase activity in vitro; this kinase activity is required for its function. BSK1 physically associates with the PAMP receptor FLAGELLIN SENSING2 and is required for a subset of flg22-induced responses, including the reactive oxygen burst, but not for mitogen-activated protein kinase activation. Our data demonstrate that BSK1 is involved in positive regulation of PTI. Together with previous findings, our work indicates that BSK1 represents a key component directly involved in both BR signaling and plant immunity.
Figures




Similar articles
-
BRASSINOSTEROID-SIGNALING KINASE1 Phosphorylates MAPKKK5 to Regulate Immunity in Arabidopsis.Plant Physiol. 2018 Apr;176(4):2991-3002. doi: 10.1104/pp.17.01757. Epub 2018 Feb 12. Plant Physiol. 2018. PMID: 29440595 Free PMC article.
-
BSK1, a receptor-like cytoplasmic kinase, involved in both BR signaling and innate immunity in Arabidopsis.Plant Signal Behav. 2013 Aug;8(8):e24996. doi: 10.4161/psb.24996. Epub 2013 May 20. Plant Signal Behav. 2013. PMID: 23733062 Free PMC article.
-
RECEPTOR-LIKE KINASE 902 Associates with and Phosphorylates BRASSINOSTEROID-SIGNALING KINASE1 to Regulate Plant Immunity.Mol Plant. 2019 Jan 7;12(1):59-70. doi: 10.1016/j.molp.2018.10.008. Epub 2018 Nov 5. Mol Plant. 2019. PMID: 30408577
-
The Multifaceted Ubiquitination of BIK1 During Plant Immunity in Arabidopsis thaliana.Int J Mol Sci. 2024 Nov 13;25(22):12187. doi: 10.3390/ijms252212187. Int J Mol Sci. 2024. PMID: 39596247 Free PMC article. Review.
-
The cell surface is the place to be for brassinosteroid perception and responses.Nat Plants. 2024 Feb;10(2):206-218. doi: 10.1038/s41477-024-01621-2. Epub 2024 Feb 22. Nat Plants. 2024. PMID: 38388723 Review.
Cited by
-
Brassinosteroid Signaling in Plant⁻Microbe Interactions.Int J Mol Sci. 2018 Dec 17;19(12):4091. doi: 10.3390/ijms19124091. Int J Mol Sci. 2018. PMID: 30563020 Free PMC article. Review.
-
A brassinosteroid-signaling kinase interacts with multiple receptor-like kinases in Arabidopsis.Mol Plant. 2014 Feb;7(2):441-4. doi: 10.1093/mp/sst105. Epub 2013 Sep 19. Mol Plant. 2014. PMID: 24051702 Free PMC article. No abstract available.
-
Constitutive signaling activity of a receptor-associated protein links fertilization with embryonic patterning in Arabidopsis thaliana.Proc Natl Acad Sci U S A. 2019 Mar 19;116(12):5795-5804. doi: 10.1073/pnas.1815866116. Epub 2019 Mar 4. Proc Natl Acad Sci U S A. 2019. PMID: 30833400 Free PMC article.
-
Broad-spectrum resistance gene RPW8.1 balances immunity and growth via feedback regulation of WRKYs.Plant Biotechnol J. 2024 Jan;22(1):116-130. doi: 10.1111/pbi.14172. Epub 2023 Sep 26. Plant Biotechnol J. 2024. PMID: 37752622 Free PMC article.
-
Mutation of the Glucosinolate Biosynthesis Enzyme Cytochrome P450 83A1 Monooxygenase Increases Camalexin Accumulation and Powdery Mildew Resistance.Front Plant Sci. 2016 Mar 2;7:227. doi: 10.3389/fpls.2016.00227. eCollection 2016. Front Plant Sci. 2016. PMID: 26973671 Free PMC article.
References
-
- Adam L., Somerville S.C. (1996). Genetic characterization of five powdery mildew disease resistance loci in Arabidopsis thaliana. Plant J. 9: 341–356 - PubMed
-
- Albrecht C., Boutrot F., Segonzac C., Schwessinger B., Gimenez-Ibanez S., Chinchilla D., Rathjen J.P., de Vries S.C., Zipfel C. (2012). Brassinosteroids inhibit pathogen-associated molecular pattern-triggered immune signaling independent of the receptor kinase BAK1. Proc. Natl. Acad. Sci. USA 109: 303–308 - PMC - PubMed
-
- Bent A.F., Mackey D. (2007). Elicitors, effectors, and R genes: The new paradigm and a lifetime supply of questions. Annu. Rev. Phytopathol. 45: 399–436 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

