A novel automethylation reaction in the Aspergillus nidulans LaeA protein generates S-methylmethionine
- PMID: 23532849
- PMCID: PMC3656261
- DOI: 10.1074/jbc.M113.465765
A novel automethylation reaction in the Aspergillus nidulans LaeA protein generates S-methylmethionine
Abstract
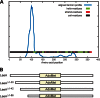
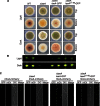
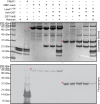
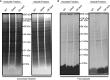
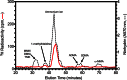
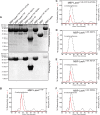
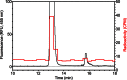
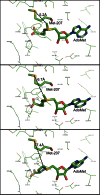
The filamentous fungi in the genus Aspergillus are opportunistic plant and animal pathogens that can adapt to their environment by producing various secondary metabolites, including lovastatin, penicillin, and aflatoxin. The synthesis of these small molecules is dependent on gene clusters that are globally regulated by the LaeA protein. Null mutants of LaeA in all pathogenic fungi examined to date show decreased virulence coupled with reduced secondary metabolism. Although the amino acid sequence of LaeA contains the motifs characteristic of seven-β-strand methyltransferases, a methyl-accepting substrate of LaeA has not been identified. In this work we did not find a methyl-accepting substrate in Aspergillus nidulans with various assays, including in vivo S-adenosyl-[methyl-(3)H]methionine labeling, targeted in vitro methylation experiments using putative protein substrates, or in vitro methylation assays using whole cell extracts grown under different conditions. However, in each experiment LaeA was shown to self-methylate. Amino acid hydrolysis of radioactively labeled LaeA followed by cation exchange and reverse phase chromatography identified methionine as the modified residue. Point mutations show that the major site of modification of LaeA is on methionine 207. However, in vivo complementation showed that methionine 207 is not required for the biological function of LaeA. LaeA is the first protein to exhibit automethylation at a methionine residue. These findings not only indicate LaeA may perform novel chemistry with S-adenosylmethionine but also provide new insights into the physiological function of LaeA.
Keywords: Aspergillus; Automethylation; Gene Regulation; Histone Methylation; LaeA; Methionine; Protein Methylation; S-Adenosylmethionine; S-Methylmethionine; Secondary Metabolism.
Figures




References
-
- Latgé J. P. (2001) The pathobiology of Aspergillus fumigatus. Trends Microbiol. 9, 382–389 - PubMed
-
- Alberts A. W., Chen J., Kuron G., Hunt V., Huff J., Hoffman C., Rothrock J., Lopez M., Joshua H., Harris E., Patchett A., Monaghan R., Currie S., Stapley E., Albers-Schonberg G., Hensens O., Hirshfield J., Hoogsteen K., Liesch J., Springer J. (1980) Mevinolin: a highly potent competitive inhibitor of hydroxymethylglutaryl-coenzyme A reductase and a cholesterol-lowering agent. Proc. Natl. Acad. Sci. U.S.A. 77, 3957–3961 - PMC - PubMed
-
- Keller N. P., Turner G., Bennett J. W. (2005) Fungal secondary metabolism–from biochemistry to genomics. Nat. Rev. Microbiol. 3, 937–947 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

