Cis-regulatory sequence variation and association with Mycoplasma load in natural populations of the house finch (Carpodacus mexicanus)
- PMID: 23532859
- PMCID: PMC3605853
- DOI: 10.1002/ece3.484
Cis-regulatory sequence variation and association with Mycoplasma load in natural populations of the house finch (Carpodacus mexicanus)
Abstract
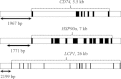
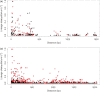
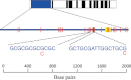
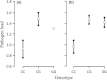
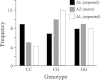
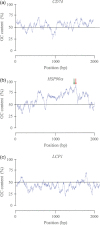
Characterization of the genetic basis of fitness traits in natural populations is important for understanding how organisms adapt to the changing environment and to novel events, such as epizootics. However, candidate fitness-influencing loci, such as regulatory regions, are usually unavailable in nonmodel species. Here, we analyze sequence data from targeted resequencing of the cis-regulatory regions of three candidate genes for disease resistance (CD74, HSP90α, and LCP1) in populations of the house finch (Carpodacus mexicanus) historically exposed (Alabama) and naïve (Arizona) to Mycoplasma gallisepticum. Our study, the first to quantify variation in regulatory regions in wild birds, reveals that the upstream regions of CD74 and HSP90α are GC-rich, with the former exhibiting unusually low sequence variation for this species. We identified two SNPs, located in a GC-rich region immediately upstream of an inferred promoter site in the gene HSP90α, that were significantly associated with Mycoplasma pathogen load in the two populations. The SNPs are closely linked and situated in potential regulatory sequences: one in a binding site for the transcription factor nuclear NFYα and the other in a dinucleotide microsatellite ((GC)6). The genotype associated with pathogen load in the putative NFYα binding site was significantly overrepresented in the Alabama birds. However, we did not see strong effects of selection at this SNP, perhaps because selection has acted on standing genetic variation over an extremely short time in a highly recombining region. Our study is a useful starting point to explore functional relationships between sequence polymorphisms, gene expression, and phenotypic traits, such as pathogen resistance that affect fitness in the wild.
Keywords: Association mapping; Mycoplasma gallisepticum; cis-regulatory element; expression; house finch.
Figures
Similar articles
-
A house finch (Haemorhous mexicanus) spleen transcriptome reveals intra- and interspecific patterns of gene expression, alternative splicing and genetic diversity in passerines.BMC Genomics. 2014 Apr 24;15:305. doi: 10.1186/1471-2164-15-305. BMC Genomics. 2014. PMID: 24758272 Free PMC article.
-
House finch populations differ in early inflammatory signaling and pathogen tolerance at the peak of Mycoplasma gallisepticum infection.Am Nat. 2013 May;181(5):674-89. doi: 10.1086/670024. Epub 2013 Mar 14. Am Nat. 2013. PMID: 23594550
-
Molecular variability of house finch Mycoplasma gallisepticum isolates as revealed by sequencing and restriction fragment length polymorphism analysis of the pvpA gene.Avian Dis. 2003 Jul-Sep;47(3):640-8. doi: 10.1637/6095. Avian Dis. 2003. PMID: 14562892
-
Susceptibility of a naïve population of house finches to Mycoplasma gallisepticum.J Wildl Dis. 2002 Apr;38(2):282-6. doi: 10.7589/0090-3558-38.2.282. J Wildl Dis. 2002. PMID: 12038126
-
Immune responses of wild birds to emerging infectious diseases.Parasite Immunol. 2015 May;37(5):242-54. doi: 10.1111/pim.12191. Parasite Immunol. 2015. PMID: 25847450 Review.
Cited by
-
Fitness consequences of structural variation inferred from a House Finch pangenome.Proc Natl Acad Sci U S A. 2024 Nov 19;121(47):e2409943121. doi: 10.1073/pnas.2409943121. Epub 2024 Nov 12. Proc Natl Acad Sci U S A. 2024. PMID: 39531493 Free PMC article.
-
Cis-regulatory evolution in a wild primate: Infection-associated genetic variation drives differential expression of MHC-DQA1 in vitro.Mol Ecol. 2017 Sep;26(17):4523-4535. doi: 10.1111/mec.14221. Epub 2017 Jul 24. Mol Ecol. 2017. PMID: 28665019 Free PMC article.
-
Understanding the evolution of immune genes in jawed vertebrates.J Evol Biol. 2023 Jun;36(6):847-873. doi: 10.1111/jeb.14181. Epub 2023 May 31. J Evol Biol. 2023. PMID: 37255207 Free PMC article. Review.
-
SNPs across time and space: population genomic signatures of founder events and epizootics in the House Finch (Haemorhous mexicanus).Ecol Evol. 2016 Sep 28;6(20):7475-7489. doi: 10.1002/ece3.2444. eCollection 2016 Oct. Ecol Evol. 2016. PMID: 28725414 Free PMC article.
-
A house finch (Haemorhous mexicanus) spleen transcriptome reveals intra- and interspecific patterns of gene expression, alternative splicing and genetic diversity in passerines.BMC Genomics. 2014 Apr 24;15:305. doi: 10.1186/1471-2164-15-305. BMC Genomics. 2014. PMID: 24758272 Free PMC article.
References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
-
- Bachmann HS, Siffert W, Frey UH. Successful amplification of extremely GC-rich promoter regions using a novel ‘slowdown PCR’ technique. Pharmacogenetics. 2003;13:759–766. - PubMed
-
- Backström N, Fagerberg S, Ellegren H. Genomics of natural bird populations: a gene-based set of reference markers evenly spread across the avian genome. Mol. Ecol. 2008;17:964–980. - PubMed
-
- Backström N, Zhang Q, Edwards SV. Comparative genomics in birds: evidence for adaptive evolution in passerines from a house finch (Carpodacus mexicanus) transcriptome. 2012. In Review.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

