Point Mutations in the folP Gene Partly Explain Sulfonamide Resistance of Streptococcus mutans
- PMID: 23533419
- PMCID: PMC3596926
- DOI: 10.1155/2013/367021
Point Mutations in the folP Gene Partly Explain Sulfonamide Resistance of Streptococcus mutans
Abstract
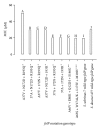
Cotrimoxazole inhibits dhfr and dhps and reportedly selects for drug resistance in pathogens. Here, Streptococcus mutans isolates were obtained from saliva of HIV/AIDS patients taking cotrimoxazole prophylaxis in Uganda. The isolates were tested for resistance to cotrimoxazole and their folP DNA (which encodes sulfonamide-targeted enzyme dhps) cloned in pUC19. A set of recombinant plasmids carrying different point mutations in cloned folP were separately transformed into folP-deficient Escherichia coli. Using sulfonamide-containing media, we assessed the growth of folP-deficient bacteria harbouring plasmids with differing folP point mutations. Interestingly, cloned folP with three mutations (A37V, N172D, R193Q) derived from Streptococcus mutans 8 conferred substantial resistance against sulfonamide to folP-deficient bacteria. Indeed, change of any of the three residues (A37V, N172D, and R193Q) in plasmid-encoded folP diminished the bacterial resistance to sulfonamide while removal of all three mutations abolished the resistance. In contrast, plasmids carrying four other mutations (A46V, E80K, Q122H, and S146G) in folP did not similarly confer any sulfonamide resistance to folP-knockout bacteria. Nevertheless, sulfonamide resistance (MIC = 50 μ M) of folP-knockout bacteria transformed with plasmid-encoded folP was much less than the resistance (MIC = 4 mM) expressed by chromosomally-encoded folP. Therefore, folP point mutations only partially explain bacterial resistance to sulfonamide.
Figures
Similar articles
-
Cotrimoxazole Prophylaxis Specifically Selects for Cotrimoxazole Resistance in Streptococcus mutans and Streptococcus sobrinus with Varied Polymorphisms in the Target Genes folA and folP.Int J Microbiol. 2012;2012:916129. doi: 10.1155/2012/916129. Epub 2012 Jan 24. Int J Microbiol. 2012. PMID: 22315614 Free PMC article.
-
Origin of the Mobile Di-Hydro-Pteroate Synthase Gene Determining Sulfonamide Resistance in Clinical Isolates.Front Microbiol. 2019 Jan 10;9:3332. doi: 10.3389/fmicb.2018.03332. eCollection 2018. Front Microbiol. 2019. PMID: 30687297 Free PMC article.
-
Sulfonamide resistance in Haemophilus influenzae mediated by acquisition of sul2 or a short insertion in chromosomal folP.Antimicrob Agents Chemother. 2002 Jun;46(6):1934-9. doi: 10.1128/AAC.46.6.1934-1939.2002. Antimicrob Agents Chemother. 2002. PMID: 12019111 Free PMC article.
-
Sulfonamide resistance in clinical isolates of Campylobacter jejuni: mutational changes in the chromosomal dihydropteroate synthase.Antimicrob Agents Chemother. 1999 Sep;43(9):2156-60. doi: 10.1128/AAC.43.9.2156. Antimicrob Agents Chemother. 1999. PMID: 10471557 Free PMC article.
-
Genetic basis for sulfonamide resistance in Bacillus anthracis.Microb Drug Resist. 2007 Spring;13(1):11-20. doi: 10.1089/mdr.2006.9992. Microb Drug Resist. 2007. PMID: 17536929
Cited by
-
Evolution of Resistance against Ciprofloxacin, Tobramycin, and Trimethoprim/Sulfamethoxazole in the Environmental Opportunistic Pathogen Stenotrophomonas maltophilia.Antibiotics (Basel). 2024 Apr 5;13(4):330. doi: 10.3390/antibiotics13040330. Antibiotics (Basel). 2024. PMID: 38667006 Free PMC article.
-
Deciphering the distance to antibiotic resistance for the pneumococcus using genome sequencing data.Sci Rep. 2017 Feb 16;7:42808. doi: 10.1038/srep42808. Sci Rep. 2017. PMID: 28205635 Free PMC article.
-
Structure-based analysis of Bacilli and plasmid dihydrofolate reductase evolution.J Mol Graph Model. 2017 Jan;71:135-153. doi: 10.1016/j.jmgm.2016.10.011. Epub 2016 Nov 22. J Mol Graph Model. 2017. PMID: 27914300 Free PMC article.
-
Whole Genome Sequencing and Characterization of Multidrug-Resistant (MDR) Bacterial Strains Isolated From a Norwegian University Campus Pond.Front Microbiol. 2020 Jun 17;11:1273. doi: 10.3389/fmicb.2020.01273. eCollection 2020. Front Microbiol. 2020. PMID: 32625184 Free PMC article.
References
-
- Bryskier A. Viridans group streptococci: a reservoir of resistant bacteria in oral cavities. Clinical Microbiology and Infection. 2002;8(2):65–69. - PubMed
-
- Wilén M, Buwembo W, Sendagire H, Kironde F, Swedberg G. Cotrimoxazole resistance of Streptococcus pneumoniae and commensal streptococci from Kampala, Uganda. Scandinavian Journal of Infectious Diseases. 2009;41(2):113–121. - PubMed
-
- Echave P, Bille J, Audet C, Talla I, Vaudaux B, Gehri M. Percentage, bacterial etiology and antibiotic susceptibility of acute respiratory infection and pneumonia among children in rural Senegal. Journal of Tropical Pediatrics. 2003;49(1):28–32. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

