The α crystallin domain of small heat shock protein b8 (Hspb8) acts as survival and differentiation factor in adult hippocampal neurogenesis
- PMID: 23536091
- PMCID: PMC6705050
- DOI: 10.1523/JNEUROSCI.6452-11.2013
The α crystallin domain of small heat shock protein b8 (Hspb8) acts as survival and differentiation factor in adult hippocampal neurogenesis
Abstract
Adult hippocampal neurogenesis is to a large degree controlled at the level of cell survival, and a number of potential mediators of this effect have been postulated. Here, we investigated the small heat shock protein Hspb8, which, because of its pleiotropic prosurvival effects in other systems, was considered a particularly promising candidate factor. Hspb8 is, for example, found in plaques of Alzheimer disease but exerts neuroprotective effects. We found that expression of Hspb8 increased during differentiation in vitro and was particularly associated with later stages (48-96 h) of differentiation. Gain-of-function and loss-of-function experiments supported the hypothesis that Hspb8 regulates cell survival of new neurons in vitro. In the dentate gyrus of adult mice in vivo, lentiviral overexpression of Hspb8 doubled the surviving cells and concomitantly promoted differentiation and net neurogenesis without affecting precursor cell proliferation. We also discovered that the truncated form of the crystallin domain of Hspb8 was sufficient to affect cell survival and neuronal differentiation in vitro and in vivo. Precursor cell experiments in vitro revealed that Hspb8 increases the phosphorylation of Akt and suggested that the prosurvival effect can be produced by a cell-autonomous mechanism. Analysis of hippocampal Hspb8 expression in mice of 69 strains of the recombinant inbred set BXD revealed that Hspb8 is a cis-acting gene whose expression was associated with clusters of transcript enriched in genes linked to growth factor signaling and apoptosis. Our results strongly suggest that Hspb8 and its α-crystallin domain might act as pleiotropic prosurvival factor in the adult hippocampus.
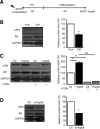
Figures










References
-
- Arndt V, Dick N, Tawo R, Dreiseidler M, Wenzel D, Hesse M, Fürst DO, Saftig P, Saint R, Fleischmann BK, Hoch M, Höhfeld J. Chaperone-assisted selective autophagy is essential for muscle maintenance. Curr Biol. 2010;20:143–148. - PubMed
-
- Bracko O, Singer T, Aigner S, Knobloch M, Winner B, Ray J, Clemenson GD, Jr, Suh H, Couillard-Despres S, Aigner L, Gage FH, Jessberger S. Gene expression profiling of neural stem cells and their neuronal progeny reveals IGF2 as a regulator of adult hippocampal neurogenesis. J Neurosci. 2012;10:3376–3387. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
