Eosinophil adoptive transfer system to directly evaluate pulmonary eosinophil trafficking in vivo
- PMID: 23536294
- PMCID: PMC3625276
- DOI: 10.1073/pnas.1220572110
Eosinophil adoptive transfer system to directly evaluate pulmonary eosinophil trafficking in vivo
Abstract
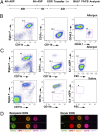
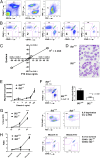
Most in vivo studies of granulocytes draw conclusions about their trafficking based on examination of their steady-state tissue/blood levels, which result from a combination of tissue homing, survival, and egress, rather than direct examination of cellular trafficking. Herein, we developed a unique cell transfer system involving the adoptive transfer of a genetically labeled, bone-marrow-derived unique granulocyte population (eosinophils) into an elicited inflammatory site, the allergic lung. A dual polychromatic FACS-based biomarker-labeling system based on the IL4-eGFP transgene (4get) or Cd45.1 allele was used to track i.v. transferred eosinophils into the airway following allergen or T(H)2-associated stimuli in the lung in multiple mouse strains. The system was amenable to reverse tagging of recipients, thus allowing transfer of nonlabeled eosinophils and competitive tracking of multiple populations of eosinophils in vivo. The half-life of eosinophils in the blood was 3 h, and migration to the lung was dependent upon the dosage of transferred eosinophils, sensitive to pertussis toxin pretreatment, peaked at ∼24 h after adoptive transfer, and revealed a greater than 8-d eosinophil half-life in the lung. Eosinophil migration to the lung was dependent upon recipient IL-5 and IL-13 receptor α1 and donor eosinophil C-C chemokine receptor type 3 (CCR3) and interleukin 1 receptor-like 1 (ST2) in vivo. Taken together, this unique eosinophil transfer system provides an unprecedented opportunity to examine airway eosinophil migration without the need for extensive efforts to acquire donor source and time-consuming genetic crossing and has already been used to identify a long eosinophil half-life in the allergic lung and a definite role for ST2 in regulating eosinophil trafficking.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Rothenberg ME, Hogan SP. The eosinophil. Annu Rev Immunol. 2006;24:147–174. - PubMed
-
- Walsh ER, Stokes K, August A. The role of eosinophils in allergic airway inflammation. Discov Med. 2010;9(47):357–362. - PubMed
-
- Shen HH, et al. A causative relationship exists between eosinophils and the development of allergic pulmonary pathologies in the mouse. J Immunol. 2003;170(6):3296–3305. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

