Quantitative PCR for detection of Shigella improves ascertainment of Shigella burden in children with moderate-to-severe diarrhea in low-income countries
- PMID: 23536399
- PMCID: PMC3716050
- DOI: 10.1128/JCM.02713-12
Quantitative PCR for detection of Shigella improves ascertainment of Shigella burden in children with moderate-to-severe diarrhea in low-income countries
Abstract
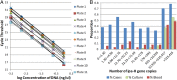
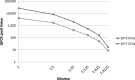
Estimates of the prevalence of Shigella spp. are limited by the suboptimal sensitivity of current diagnostic and surveillance methods. We used a quantitative PCR (qPCR) assay to detect Shigella in the stool samples of 3,533 children aged <59 months from the Gambia, Mali, Kenya, and Bangladesh, with or without moderate-to-severe diarrhea (MSD). We compared the results from conventional culture to those from qPCR for the Shigella ipaH gene. Using MSD as the reference standard, we determined the optimal cutpoint to be 2.9 × 10(4) ipaH copies per 100 ng of stool DNA for set 1 (n = 877). One hundred fifty-eight (18%) specimens yielded >2.9 × 10(4) ipaH copies. Ninety (10%) specimens were positive by traditional culture for Shigella. Individuals with ≥ 2.9 × 10(4) ipaH copies have 5.6-times-higher odds of having diarrhea than those with <2.9 × 10(4) ipaH copies (95% confidence interval, 3.7 to 8.5; P < 0.0001). Nearly identical results were found using an independent set of samples. qPCR detected 155 additional MSD cases with high copy numbers of ipaH, a 90% increase from the 172 cases detected by culture in both samples. Among a subset (n = 2,874) comprising MSD cases and their age-, gender-, and location-matched controls, the fraction of MSD cases that were attributable to Shigella infection increased from 9.6% (n = 129) for culture to 17.6% (n = 262) for qPCR when employing our cutpoint. We suggest that qPCR with a cutpoint of approximately 1.4 × 10(4) ipaH copies be the new reference standard for the detection and diagnosis of shigellosis in children in low-income countries. The acceptance of this new standard would substantially increase the fraction of MSD cases that are attributable to Shigella.
Figures

References
-
- von Seidlein L, Kim DR, Ali M, Lee H, Wang X, Thiem VD, Canh do G, Chaicumpa W, Agtini MD, Hossain A, Bhutta ZA, Mason C, Sethabutr O, Talukder K, Nair GB, Deen JL, Kotloff K, Clemens J. 2006. A multicentre study of Shigella diarrhoea in six Asian countries: disease burden, clinical manifestations, and microbiology. PLoS Med. 3:e353 doi:10.1371/journal.pmed.0030353 - DOI - PMC - PubMed
-
- Taniuchi M, Walters CC, Gratz J, Maro A, Kumburu H, Serichantalergs O, Sethabutr O, Bodhidatta L, Kibiki G, Toney DM, Berkeley L, Nataro JP, Houpt ER. 2012. Development of a multiplex polymerase chain reaction assay for diarrheagenic Escherichia coli and Shigella spp. and its evaluation on colonies, culture broths, and stool. Diagn. Microbiol. Infect. Dis. 73:121–128 - PMC - PubMed
-
- Vu DT, Sethabutr O, Von Seidlein L, Tran VT, Do GC, Bui TC, Le HT, Lee H, Houng HS, Hale TL, Clemens JD, Mason C, Dang DT. 2004. Detection of Shigella by a PCR assay targeting the ipaH gene suggests increased prevalence of shigellosis in Nha Trang, Vietnam. J. Clin. Microbiol. 42:2031–2035 - PMC - PubMed
-
- Samosornsuk S, Chaicumpa W, von Seidlein L, Clemens JD, Sethabutr O. 2007. Using real-time PCR to detect shigellosis: ipaH detection in Kaeng-Khoi District, Saraburi Province, Thailand. Thammasat Int. J. Sci. Technol. 12:52–57
-
- Phantouamath B, Sithivong N, Insisiengmay S, Ichinose Y, Higa N, Song T, Iwanaga M. 2005. Pathogenicity of Shigella in healthy carriers: a study in Vientiane, Lao People's Democratic Republic. Jpn. J. Infect. Dis. 58:232–234 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

