TMPRSS2 activates the human coronavirus 229E for cathepsin-independent host cell entry and is expressed in viral target cells in the respiratory epithelium
- PMID: 23536651
- PMCID: PMC3648130
- DOI: 10.1128/JVI.03372-12
TMPRSS2 activates the human coronavirus 229E for cathepsin-independent host cell entry and is expressed in viral target cells in the respiratory epithelium
Abstract
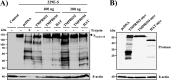
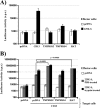
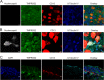
Infection with human coronavirus 229E (HCoV-229E) is associated with the common cold and may result in pneumonia in immunocompromised patients. The viral spike (S) protein is incorporated into the viral envelope and mediates infectious entry of HCoV-229E into host cells, a process that depends on the activation of the S-protein by host cell proteases. However, the proteases responsible for HCoV-229E activation are incompletely defined. Here we show that the type II transmembrane serine proteases TMPRSS2 and HAT cleave the HCoV-229E S-protein (229E-S) and augment 229E-S-driven cell-cell fusion, suggesting that TMPRSS2 and HAT can activate 229E-S. Indeed, engineered expression of TMPRSS2 and HAT rendered 229E-S-driven virus-cell fusion insensitive to an inhibitor of cathepsin L, a protease previously shown to facilitate HCoV-229E infection. Inhibition of endogenous cathepsin L or TMPRSS2 demonstrated that both proteases can activate 229E-S for entry into cells that are naturally susceptible to infection. In addition, evidence was obtained that activation by TMPRSS2 rescues 229E-S-dependent cell entry from inhibition by IFITM proteins. Finally, immunohistochemistry revealed that TMPRSS2 is coexpressed with CD13, the HCoV-229E receptor, in human airway epithelial (HAE) cells, and that CD13(+) TMPRSS2(+) cells are preferentially targeted by HCoV-229E, suggesting that TMPRSS2 can activate HCoV-229E in infected humans. In sum, our results indicate that HCoV-229E can employ redundant proteolytic pathways to ensure its activation in host cells. In addition, our observations and previous work suggest that diverse human respiratory viruses are activated by TMPRSS2, which may constitute a target for antiviral intervention.
Figures



References
-
- Bermingham A, Chand M, Brown C, Aarons E, Tong C, Langrish C, Hoschler K, Brown K, Galiano M, Myers R, Pebody R, Green H, Boddington N, Gopal R, Price N, Newsholme W, Drosten C, Fouchier R, Zambon M. 2012. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro Surveill. 17:20290. - PubMed
-
- Corman V, Eckerle I, Bleicker T, Zaki A, Landt O, Eschbach-Bludau M, BSvan Gopal R, Ballhause M, Bestebroer T, Muth D, Muller M, Drexler J, Zambon M, Osterhaus A, Fouchier R, Drosten C. 2012. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill. 17:20285. - PubMed
-
- Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RA, Berger A, Burguiere AM, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra JC, Muller S, Rickerts V, Sturmer M, Vieth S, Klenk HD, Osterhaus AD, Schmitz H, Doerr HW. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967–1976 - PubMed
-
- Ksiazek TG, Erdman D, Goldsmith CS, Zaki SR, Peret T, Emery S, Tong S, Urbani C, Comer JA, Lim W, Rollin PE, Dowell SF, Ling AE, Humphrey CD, Shieh WJ, Guarner J, Paddock CD, Rota P, Fields B, DeRisi J, Yang JY, Cox N, Hughes JM, LeDuc JW, Bellini WJ, Anderson LJ. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953–1966 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

