Tyrosine 132 phosphorylation of influenza A virus M1 protein is crucial for virus replication by controlling the nuclear import of M1
- PMID: 23536660
- PMCID: PMC3648105
- DOI: 10.1128/JVI.03024-12
Tyrosine 132 phosphorylation of influenza A virus M1 protein is crucial for virus replication by controlling the nuclear import of M1
Abstract
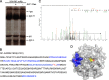
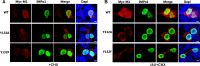
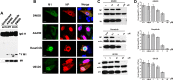
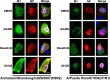
Phosphorylation of viral proteins plays important roles in the influenza A virus (IAV) life cycle. By using mass spectrometry, we identified tyrosine 132 (Y132) as a phosphorylation site of the matrix protein (M1) of the influenza virus A/WSN/1933(H1N1). Phosphorylation at this site is essential to the process of virus replication by controlling the nuclear import of M1. We further demonstrated that the phosphorylated tyrosine is crucial for the binding of M1 to the nuclear import factor importin-α1, since any substitutions at this site severely reduce this protein-protein interaction and damage the importin-α1-mediated nuclear import of M1. Additionally, the tyrosine phosphorylation which leads to the nuclear import of M1 is blocked by a Janus kinase inhibitor. The present study reveals a pivotal role of this tyrosine phosphorylation in the intracellular transportation of M1, which controls the process of viral replication.
Figures





References
-
- Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, Dalton RM, Lo A, Efstathiou S, Atkins JF, Firth AE, Taubenberger JK, Digard P. 2012. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 337:199–204 - PMC - PubMed
-
- Wise HM, Hutchinson EC, Jagger BW, Stuart AD, Kang ZH, Robb N, Schwartzman LM, Kash JC, Fodor E, Firth AE, Gog JR, Taubenberger JK, Digard P. 2012. Identification of a novel splice variant form of the influenza A virus M2 ion channel with an antigenically distinct ectodomain. PLoS Pathog. 8:e1002998 doi:10.1371/journal.ppat.1002998 - DOI - PMC - PubMed
-
- Gregoriades A, Guzman GG, Paoletti E. 1990. The phosphorylation of the integral membrane (M1) protein of influenza virus. Virus Res. 16:27–41 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

