HIV-1 Gag associates with specific uropod-directed microdomains in a manner dependent on its MA highly basic region
- PMID: 23536680
- PMCID: PMC3648103
- DOI: 10.1128/JVI.00040-13
HIV-1 Gag associates with specific uropod-directed microdomains in a manner dependent on its MA highly basic region
Abstract
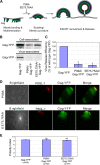
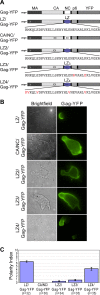
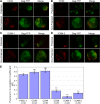
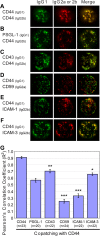
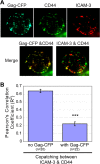
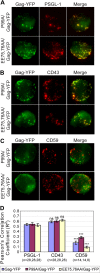
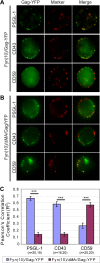
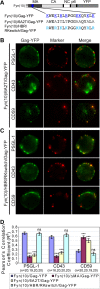
In polarized T cells, HIV-1 Gag localizes to a rear-end protrusion known as the uropod in a multimerization-dependent manner. Gag-laden uropods participate in formation of virological synapses, intercellular contact structures that play a key role in cell-to-cell HIV-1 transmission. Our previous observations suggest that Gag associates with uropod-directed microdomains (UDMs) that eventually comigrate with Gag to the uropod over the cell surface. However, the nature of Gag multimerization required for this movement, the composition of the UDMs, and the molecular determinants for Gag association with these microdomains remain unknown. In this study, we found that Gag multimerization prior to budding but beyond dimerization is necessary for Gag localization to the uropods, indicating that uropod localization occurs early in the assembly process. We also found that prior to membrane curvature, Gag multimers associate with a specific subset of UDMs containing PSGL-1, CD43, and CD44 but not ICAM-1, ICAM-3, or CD59. Notably, upon association, Gag excludes ICAM-3 from this subset of UDMs, revealing an active and selective reorganization of these microdomains by Gag. This specific association between Gag and UDMs is dependent on the highly basic region (HBR) in the Gag matrix (MA) domain. The overall positive charge of the HBR was needed for the interaction with the specific UDM subset, while the exact HBR sequence was not, unlike that seen for MA binding to the plasma membrane phospholipid phosphatidylinositol-(4,5)-bisphosphate [PI(4,5)P2]. Taken together, these findings revealed that HIV-1 Gag associates with specific microdomains present in polarized T cells in an MA-dependent manner, which results in modification of the microdomain constituents.
Figures

References
-
- Mazurov D, Ilinskaya A, Heidecker G, Lloyd P, Derse D. 2010. Quantitative comparison of HTLV-1 and HIV-1 cell-to-cell infection with new replication dependent vectors. PLoS Pathog. 6:e1000788 doi:10.1371/journal.ppat.1000788 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

