Identification and analysis of the acetylated status of poplar proteins reveals analogous N-terminal protein processing mechanisms with other eukaryotes
- PMID: 23536812
- PMCID: PMC3594182
- DOI: 10.1371/journal.pone.0058681
Identification and analysis of the acetylated status of poplar proteins reveals analogous N-terminal protein processing mechanisms with other eukaryotes
Abstract
Background: The N-terminal protein processing mechanism (NPM) including N-terminal Met excision (NME) and N-terminal acetylation (N(α)-acetylation) represents a common protein co-translational process of some eukaryotes. However, this NPM occurred in woody plants yet remains unknown.
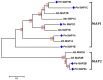
Methodology/principal findings: To reveal the NPM in poplar, we investigated the N(α)-acetylation status of poplar proteins during dormancy by combining tandem mass spectrometry with TiO2 enrichment of acetylated peptides. We identified 58 N-terminally acetylated (N(α)-acetylated) proteins. Most proteins (47, >81%) are subjected to N(α)-acetylation following the N-terminal removal of Met, indicating that N(α)-acetylation and NME represent a common NPM of poplar proteins. Furthermore, we confirm that poplar shares the analogous NME and N(α)-acetylation (NPM) to other eukaryotes according to analysis of N-terminal features of these acetylated proteins combined with genome-wide identification of the involving methionine aminopeptidases (MAPs) and N-terminal acetyltransferase (Nat) enzymes in poplar. The N(α)-acetylated reactions and the involving enzymes of these poplar proteins are also identified based on those of yeast and human, as well as the subcellular location information of these poplar proteins.
Conclusions/significance: This study represents the first extensive investigation of N(α)-acetylation events in woody plants, the results of which will provide useful resources for future unraveling the regulatory mechanisms of N(α)-acetylation of proteins in poplar.
Conflict of interest statement
Figures
Similar articles
-
In silico identification and characterization of N-Terminal acetyltransferase genes of poplar (Populus trichocarpa).Int J Mol Sci. 2014 Jan 27;15(2):1852-64. doi: 10.3390/ijms15021852. Int J Mol Sci. 2014. PMID: 24473137 Free PMC article.
-
Genome-wide identification and in silico analysis of poplar peptide deformylases.Int J Mol Sci. 2012;13(4):5112-5124. doi: 10.3390/ijms13045112. Epub 2012 Apr 23. Int J Mol Sci. 2012. PMID: 22606033 Free PMC article.
-
Characterization of N-terminal protein modifications in Pseudomonas aeruginosa PA14.J Proteomics. 2015 Jan 30;114:214-25. doi: 10.1016/j.jprot.2014.11.006. Epub 2014 Nov 21. J Proteomics. 2015. PMID: 25464366
-
N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins.J Mol Biol. 2003 Jan 24;325(4):595-622. doi: 10.1016/s0022-2836(02)01269-x. J Mol Biol. 2003. PMID: 12507466 Review.
-
Co-translational, Post-translational, and Non-catalytic Roles of N-Terminal Acetyltransferases.Mol Cell. 2019 Mar 21;73(6):1097-1114. doi: 10.1016/j.molcel.2019.02.007. Epub 2019 Mar 13. Mol Cell. 2019. PMID: 30878283 Free PMC article. Review.
Cited by
-
Transcriptome Sequencing of Lima Bean (Phaseolus lunatus) to Identify Putative Positive Selection in Phaseolus and Legumes.Int J Mol Sci. 2015 Jul 3;16(7):15172-87. doi: 10.3390/ijms160715172. Int J Mol Sci. 2015. PMID: 26151849 Free PMC article.
-
Two N-terminal acetyltransferases antagonistically regulate the stability of a nod-like receptor in Arabidopsis.Plant Cell. 2015 May;27(5):1547-62. doi: 10.1105/tpc.15.00173. Epub 2015 May 12. Plant Cell. 2015. PMID: 25966763 Free PMC article.
-
The biological functions of Naa10 - From amino-terminal acetylation to human disease.Gene. 2015 Aug 10;567(2):103-31. doi: 10.1016/j.gene.2015.04.085. Epub 2015 May 16. Gene. 2015. PMID: 25987439 Free PMC article. Review.
-
Dual lysine and N-terminal acetyltransferases reveal the complexity underpinning protein acetylation.Mol Syst Biol. 2020 Jul;16(7):e9464. doi: 10.15252/msb.20209464. Mol Syst Biol. 2020. PMID: 32633465 Free PMC article.
-
In silico identification and characterization of N-Terminal acetyltransferase genes of poplar (Populus trichocarpa).Int J Mol Sci. 2014 Jan 27;15(2):1852-64. doi: 10.3390/ijms15021852. Int J Mol Sci. 2014. PMID: 24473137 Free PMC article.
References
-
- Polevoda B, Sherman F (2003) N-terminal acetyltransferases and sequence requirements for N-terminal acetylation of eukaryotic proteins. Journal of Molecular Biology 325: 595–622. - PubMed
-
- Martinez A, Traverso JA, Valot B, Ferro M, Espagne C, et al. (2008) Extent of N-terminal modifications in cytosolic proteins from eukaryotes. Proteomics 8: 2809–2831. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

