Characterization and functional analysis of the potato pollen-specific microtubule-associated protein SBgLR in tobacco
- PMID: 23536914
- PMCID: PMC3607588
- DOI: 10.1371/journal.pone.0060543
Characterization and functional analysis of the potato pollen-specific microtubule-associated protein SBgLR in tobacco
Abstract
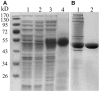
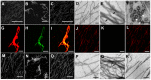
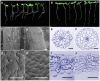
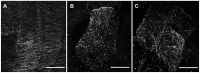
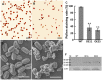
Microtubule-associated proteins play a crucial role in the regulation of microtubule dynamics, and are very important for plant cell and organ development. SBgLR is a potato pollen-specific protein, with five imperfect V-V-E-K-K-N/E-E repetitive motifs that are responsible for microtubule binding activity. In present study, SBgLR showed typical microtubule-associated protein characteristics; it bound tubulin and microtubules, and colocalized with microtubules in vitro. We also found that SBgLR could form oligomers, and that both the SBgLR monomers and oligomers bundle microtubules in vitro. Constitutive expression of SBgLR in tobacco caused curving and right-handed twisting root growth, abnormal directional cell expansion and cell layer arrangement, and pollen abortion. Immunofluorescence staining assays revealed that microtubule organization is altered in root epidermal cells in SBgLR-overexpressing lines. These suggest that SBgLR functions as a microtubule-associated protein in pollen development. Our results indicate that normal organization of MTs may be crucial for pollen development.
Conflict of interest statement
Figures




References
-
- Mayer U, Jürgens G (2002) Microtubule cytoskeleton: a track record. Curr Opin Plant Biol 5: 494–501. - PubMed
-
- Smith LG (2003) Cytoskeletal control of plant cell shape: getting the fine points. Curr Opin Plant Biol 6: 63–73. - PubMed
-
- Hashimoto T (2003) Dynamics and regulation of plant interphase microtubules: a comparative view. Curr Opin Plant Biol 6: 568–576. - PubMed
-
- Lloyd C, Chan J (2004) Microtubules and the shape of plants to come. Nat Rev Mol Cell Biol 5: 13–22. - PubMed
-
- Mathur J, Hülskamp M (2002) Microtubules and microfilaments in cell morphogenesis in higher plants. Curr Biol 12: 669–676. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

