Genome sequence of ground tit Pseudopodoces humilis and its adaptation to high altitude
- PMID: 23537097
- PMCID: PMC4053790
- DOI: 10.1186/gb-2013-14-3-r29
Genome sequence of ground tit Pseudopodoces humilis and its adaptation to high altitude
Erratum in
- Genome Biol. 2014;15(2):R33. Lei, Fumin [removed]
Abstract
Background: The mechanism of high-altitude adaptation has been studied in certain mammals. However, in avian species like the ground tit Pseudopodoces humilis, the adaptation mechanism remains unclear. The phylogeny of the ground tit is also controversial.
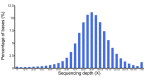
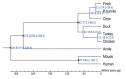
Results: Using next generation sequencing technology, we generated and assembled a draft genome sequence of the ground tit. The assembly contained 1.04 Gb of sequence that covered 95.4% of the whole genome and had higher N50 values, at the level of both scaffolds and contigs, than other sequenced avian genomes. About 1.7 million SNPs were detected, 16,998 protein-coding genes were predicted and 7% of the genome was identified as repeat sequences. Comparisons between the ground tit genome and other avian genomes revealed a conserved genome structure and confirmed the phylogeny of ground tit as not belonging to the Corvidae family. Gene family expansion and positively selected gene analysis revealed genes that were related to cardiac function. Our findings contribute to our understanding of the adaptation of this species to extreme environmental living conditions.
Conclusions: Our data and analysis contribute to the study of avian evolutionary history and provide new insights into the adaptation mechanisms to extreme conditions in animals.
Figures
References
-
- Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZX, Pool JE, Xu X, Jiang H, Vinckenbosch N, Korneliussen TS, Zheng H, Liu T, He W, Li K, Luo R, Nie X, Wu H, Zhao M, Cao H, Zou J, Shan Y, Li S, Yang Q, Ni P, Tian G, Xu J, Liu X, Jiang T, Wu R. et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;14:75–78. doi: 10.1126/science.1190371. - DOI - PMC - PubMed
-
- Beall CM, Cavalleri GL, Deng L, Elston RC, Gao Y, Knight J, Li C, Li JC, Liang Y, McCormack M, Montgomery HE, Pan H, Robbins PA, Shianna KV, Tam SC, Tsering N, Veeramah KR, Wang W, Wangdui P, Weale ME, Xu Y, Xu Z, Yang L, Zaman MJ, Zeng C, Zhang L, Zhang X, Zhaxi P, Zheng YT. Natural selection on EPAS1 (HIF2alpha) associated with low hemoglobin concentration in Tibetan highlanders. PNAS. 2010;14:11459–11464. doi: 10.1073/pnas.1002443107. - DOI - PMC - PubMed
-
- Qiu Q, Zhang G, Ma T, Qian W, Wang J, Ye Z, Cao C, Hu Q, Kim J, Larkin DM, Auvil L, Capitanu B, Ma J, Lewin HA, Qian X, Lang Y, Zhou R, Wang L, Wang K, Xia J, Liao S, Pan S, Lu X, Hou H, Wang Y, Zang X, Yin Y, Ma H, Zhang J, Wang Z. et al. The yak genome and adaptation to life at high altitude. Nat Genet. 2012;14:946–9. doi: 10.1038/ng.2343. - DOI - PubMed
-
- Bulgarella M, Peters JL, Kopuchian C, Valqui T, Wilson RE, McCracken KG. Multilocus coalescent analysis of haemoglobin differentiation between low- and high-altitude populations of crested ducks (Lophonetta specularioides). Mol Ecol. 2012;14:350–68. doi: 10.1111/j.1365-294X.2011.05400.x. - DOI - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

