Two hot spot mutant p53 mouse models display differential gain of function in tumorigenesis
- PMID: 23538418
- PMCID: PMC3679454
- DOI: 10.1038/cdd.2013.17
Two hot spot mutant p53 mouse models display differential gain of function in tumorigenesis
Erratum in
-
Correction to: Two hot spot mutant p53 mouse models display differential gain of function in tumorigenesis.Cell Death Differ. 2024 Oct;31(10):1394. doi: 10.1038/s41418-024-01366-7. Cell Death Differ. 2024. PMID: 39256520 Free PMC article. No abstract available.
Abstract
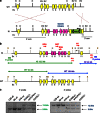
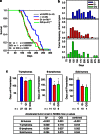
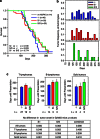
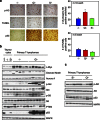
Mutant p53 proteins not only lose their tumor-suppressor function but some acquire oncogenic gain of function (GOF). The published mutp53 knock-in (KI) alleles (R172H, R270H, R248W) manifest GOF by broader tumor spectrum and more metastasis compared with the p53-null allele, but do not shorten survival. However, whether GOF also occurs with other mutations and whether they are all biologically equal is unknown. To answer this, we created novel humanized mutp53 KI mice harboring the hot spot alleles R248Q and G245S. Intriguingly, their impact was very different. Compared with p53-null mice, R248Q/- mice had accelerated onset of all tumor types and shorter survival, thus unprecedented strong GOF. In contrast, G245S/- mice were similar to null mice in tumor latency and survival. This was associated with a twofold higher T-lymphoma proliferation in R248Q/- mice compared with G245S/- and null mice. Moreover, R248Q/- hematopoietic and mesenchymal stem cells were expanded relative to G245S/- and null mice, the first indication that GOF also acts by perturbing pretumorous progenitor pools. Importantly, these models closely mirror Li-Fraumeni patients who show higher tumor numbers, accelerated onset and shorter tumor-free survival by 10.5 years when harboring codon R248Q mutations as compared with Li-Fraumeni patients with codon G245S mutations or p53 deletions/loss. Conversely, both KI alleles caused a modest broadening of tumor spectrum with enhanced Akt signaling compared with null mice. These models are the first in vivo proof for differential oncogenic strength among p53 GOF alleles, with genotype-phenotype correlations borne out in humans.
Figures



Comment in
-
Not all p53 gain-of-function mutants are created equal.Cell Death Differ. 2013 Jul;20(7):855-7. doi: 10.1038/cdd.2013.53. Cell Death Differ. 2013. PMID: 23749181 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

