Improved ancestry inference using weights from external reference panels
- PMID: 23539302
- PMCID: PMC3661048
- DOI: 10.1093/bioinformatics/btt144
Improved ancestry inference using weights from external reference panels
Abstract
Motivation: Inference of ancestry using genetic data is motivated by applications in genetic association studies, population genetics and personal genomics. Here, we provide methods and software for improved ancestry inference using genome-wide single nucleotide polymorphism (SNP) weights from external reference panels. This approach makes it possible to leverage the rich ancestry information that is available from large external reference panels, without the administrative and computational complexities of re-analyzing the raw genotype data from the reference panel in subsequent studies.
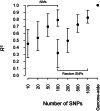
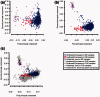
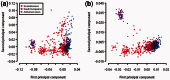
Results: We extensively validate our approach in multiple African American, Latino American and European American datasets, making use of genome-wide SNP weights derived from large reference panels, including HapMap 3 populations and 6546 European Americans from the Framingham Heart Study. We show empirically that our approach provides much greater accuracy than either the prevailing ancestry-informative marker (AIM) approach or the analysis of genome-wide target genotypes without a reference panel. For example, in an independent set of 1636 European American genome-wide association study samples, we attained prediction accuracy (R(2)) of 1.000 and 0.994 for the first two principal components using our method, compared with 0.418 and 0.407 using 150 published AIMs or 0.955 and 0.003 by applying principal component analysis directly to the target samples. We finally show that the higher accuracy in inferring ancestry using our method leads to more effective correction for population stratification in association studies.
Availability: The SNPweights software is available online at http://www.hsph.harvard.edu/faculty/alkes-price/software/.
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

