Biosynthesis of the urease metallocenter
- PMID: 23539618
- PMCID: PMC3650357
- DOI: 10.1074/jbc.R112.446526
Biosynthesis of the urease metallocenter
Abstract
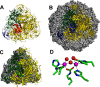
Metalloenzymes often require elaborate metallocenter assembly systems to create functional active sites. The medically important dinuclear nickel enzyme urease provides an excellent model for studying metallocenter assembly. Nickel is inserted into the urease active site in a GTP-dependent process with the assistance of UreD/UreH, UreE, UreF, and UreG. These accessory proteins orchestrate apoprotein activation by delivering the appropriate metal, facilitating protein conformational changes, and possibly providing a requisite post-translational modification. The activation mechanism and roles of each accessory protein in urease maturation are the subject of ongoing studies, with the latest findings presented in this minireview.
Keywords: Biosynthesis; Chaperone Chaperonin; GTPase; Metalloenzymes; Nickel; Urease.
Figures
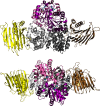

References
-
- Scott D. R., Marcus E. A., Weeks D. L., Sachs G. (2002) Mechanisms of acid resistance due to the urease system of Helicobacter pylori. Gastroenterology 123, 187–195 - PubMed
-
- Nielubowicz G. R., Mobley H. L. T. (2010) Host-pathogen interactions in the urinary tract interaction. Nat. Rev. Urol. 7, 430–441 - PubMed
-
- Witte C.-P. (2011) Urea metabolism in plants. Plant Sci. 180, 431–438 - PubMed
-
- Bremner J. M. (1995) Recent research on problems in the use of urea as a nitrogen fertilizer. Fertilizer Res. 42, 321–329
-
- Sumner J. B. (1926) The isolation and crystallization of the enzyme urease. J. Biol. Chem. 69, 435–441
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

