Modified MuDPIT separation identified 4488 proteins in a system-wide analysis of quiescence in yeast
- PMID: 23540446
- PMCID: PMC3815592
- DOI: 10.1021/pr400027m
Modified MuDPIT separation identified 4488 proteins in a system-wide analysis of quiescence in yeast
Abstract
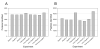
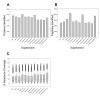
A modified multidimensional protein identification technology (MudPIT) separation was coupled to an LTQ Orbitrap Velos mass spectrometer and used to rapidly identify the near-complete yeast proteome from a whole cell tryptic digest. This modified online two-dimensional liquid chromatography separation consists of 39 strong cation exchange steps followed by a short 18.5 min reversed-phase (RP) gradient. A total of 4269 protein identifications were made from 4189 distinguishable protein families from yeast during log phase growth. The "Micro" MudPIT separation performed as well as a standard MudPIT separation in 40% less gradient time. The majority of the yeast proteome can now be routinely covered in less than a days' time with high reproducibility and sensitivity. The newly devised separation method was used to detect changes in protein expression during cellular quiescence in yeast. An enrichment in the GO annotations "oxidation reduction", "catabolic processing" and "cellular response to oxidative stress" was seen in the quiescent cellular fraction, consistent with their long-lived stress resistant phenotypes. Heterogeneity was observed in the stationary phase fraction with a less dense cell population showing reductions in KEGG pathway categories of "Ribosome" and "Proteasome", further defining the complex nature of yeast populations present during stationary phase growth. In total, 4488 distinguishable protein families were identified in all cellular conditions tested.
Figures



References
-
- de Godoy LM, Olsen J. V., Cox, J., Nielsen ML, Hubner NC, Fröhlich F, Walther TC, Mann M. Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature. 2008;455(7217):1251–4. - PubMed
-
- Peng M, Taouatas N, Cappadona S, Breukelen B, Mohammed S, Scholten A, Heck A. Protein bias in absolute protein quantitation. Nat. Methods. 2012;(6):524–5. - PubMed
-
- Picotti P, Rinner O, Stallmach R, Dautel F, Farrah T, Domon B, Wenschuh H, Aebersold R. High-throughput generation of selected reaction-monitoring assays for proteins and proteomes. Nat. Methods. 2010;(1):43–6. - PubMed
-
- Picotti P, Clement-Ziza M, Lam H, Campbell D, Schmidt A, Deutsch E, Rost H, Sun Z, Rinner O, Reiter L, Shen Q, Michaelson J, Frei A, Alberti S, Kusebauch U, Wollscheid B, Moritz R, Beyer A, Aebersold R. A complete mass-spectrometric map of the yeast proteome applied to quantitative trait analysis. Nature. 2013;(494):266–70. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

