The expanding bacterial type IV secretion lexicon
- PMID: 23542405
- PMCID: PMC3816095
- DOI: 10.1016/j.resmic.2013.03.012
The expanding bacterial type IV secretion lexicon
Abstract
The bacterial type IV secretion systems (T4SSs) comprise a biologically diverse group of translocation systems functioning to deliver DNA or protein substrates from donor to target cells generally by a mechanism dependent on establishment of direct cell-to-cell contact. Members of one T4SS subfamily, the conjugation systems, mediate the widespread and rapid dissemination of antibiotic resistance and virulence traits among bacterial pathogens. Members of a second subfamily, the effector translocators, are used by often medically-important pathogens to deliver effector proteins to eukaryotic target cells during the course of infection. Here we summarize our current understanding of the structural and functional diversity of T4SSs and of the evolutionary processes shaping this diversity. We compare mechanistic and architectural features of T4SSs from Gram-negative and -positive species. Finally, we introduce the concept of the 'minimized' T4SSs; these are systems composed of a conserved set of 5-6 subunits that are distributed among many Gram-positive and some Gram-negative species.
Keywords: ATPase; Conjugation; Pathogenesis; Pilus; Translocation; Type IV secretion.
Copyright © 2013 Institut Pasteur. Published by Elsevier Masson SAS. All rights reserved.
Figures



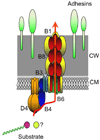

References
-
- Aly KA, Baron C. The VirB5 protein localizes to the T-pilus tips in Agrobacterium tumefaciens . Microbiology. 2007;153:3766–3775. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

