Detection and characterization of megasatellites in orthologous and nonorthologous genes of 21 fungal genomes
- PMID: 23543670
- PMCID: PMC3675995
- DOI: 10.1128/EC.00001-13
Detection and characterization of megasatellites in orthologous and nonorthologous genes of 21 fungal genomes
Abstract
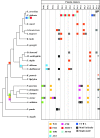
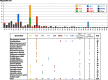
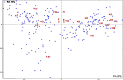
Megasatellites are large DNA tandem repeats, originally described in Candida glabrata, in protein-coding genes. Most of the genes in which megasatellites are found are of unknown function. In this work, we extended the search for megasatellites to 20 additional completely sequenced fungal genomes and extracted 216 megasatellites in 203 out of 142,121 genes, corresponding to the most exhaustive description of such genetic elements available today. We show that half of the megasatellites detected encode threonine-rich peptides predicted to be intrinsically disordered, suggesting that they may interact with several partners or serve as flexible linkers. Megasatellite motifs were clustered into several families. Their distribution in fungal genes shows that different motifs are found in orthologous genes and similar motifs are found in unrelated genes, suggesting that megasatellite formation or spreading does not necessarily track the evolution of their host genes. Altogether, these results suggest that megasatellites are created and lost during evolution of fungal genomes, probably sharing similar functions, although their primary sequences are not necessarily conserved.
Figures


Similar articles
-
Dynamic evolution of megasatellites in yeasts.Nucleic Acids Res. 2010 Aug;38(14):4731-9. doi: 10.1093/nar/gkq207. Epub 2010 Mar 31. Nucleic Acids Res. 2010. PMID: 20360043 Free PMC article.
-
Megasatellites: a peculiar class of giant minisatellites in genes involved in cell adhesion and pathogenicity in Candida glabrata.Nucleic Acids Res. 2008 Oct;36(18):5970-82. doi: 10.1093/nar/gkn594. Epub 2008 Sep 23. Nucleic Acids Res. 2008. PMID: 18812401 Free PMC article.
-
Megasatellites: a new class of large tandem repeats discovered in the pathogenic yeast Candida glabrata.Cell Mol Life Sci. 2010 Mar;67(5):671-6. doi: 10.1007/s00018-009-0216-y. Epub 2009 Nov 28. Cell Mol Life Sci. 2010. PMID: 19946728 Free PMC article. Review.
-
Loss of the flagellum happened only once in the fungal lineage: phylogenetic structure of kingdom Fungi inferred from RNA polymerase II subunit genes.BMC Evol Biol. 2006 Sep 29;6:74. doi: 10.1186/1471-2148-6-74. BMC Evol Biol. 2006. PMID: 17010206 Free PMC article.
-
Phylogeny and systematics of the fungi with special reference to the Ascomycota and Basidiomycota.Chem Immunol. 2002;81:207-95. doi: 10.1159/000058868. Chem Immunol. 2002. PMID: 12102002 Review. No abstract available.
Cited by
-
Digested disorder: Quarterly intrinsic disorder digest (April-May-June, 2013).Intrinsically Disord Proteins. 2013 Jan 1;1(1):e27454. doi: 10.4161/idp.27454. eCollection 2013 Jan-Dec. Intrinsically Disord Proteins. 2013. PMID: 28516028 Free PMC article. Review.
-
Functional variability in adhesion and flocculation of yeast megasatellite genes.Genetics. 2022 May 5;221(1):iyac042. doi: 10.1093/genetics/iyac042. Genetics. 2022. PMID: 35274698 Free PMC article.
-
Genome-wide replication landscape of Candida glabrata.BMC Biol. 2015 Sep 2;13:69. doi: 10.1186/s12915-015-0177-6. BMC Biol. 2015. PMID: 26329162 Free PMC article.
-
Genome structure and dynamics of the yeast pathogen Candida glabrata.FEMS Yeast Res. 2014 Jun;14(4):529-35. doi: 10.1111/1567-1364.12145. Epub 2014 Mar 10. FEMS Yeast Res. 2014. PMID: 24528571 Free PMC article. Review.
-
Genome Data Exploration Using Correspondence Analysis.Bioinform Biol Insights. 2016 Jun 7;10:59-72. doi: 10.4137/BBI.S39614. eCollection 2016. Bioinform Biol Insights. 2016. PMID: 27279736 Free PMC article. Review.
References
-
- Vergnaud G, Denoeud F. 2000. Minisatellites: mutability and genome architecture. Genome Res. 10:899–907 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

