Roles of N-terminal fatty acid acylations in membrane compartment partitioning: Arabidopsis h-type thioredoxins as a case study
- PMID: 23543785
- PMCID: PMC3634677
- DOI: 10.1105/tpc.112.106849
Roles of N-terminal fatty acid acylations in membrane compartment partitioning: Arabidopsis h-type thioredoxins as a case study
Abstract
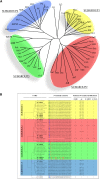
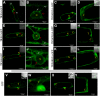
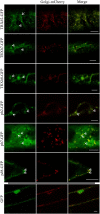
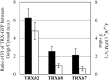
N-terminal fatty acylations (N-myristoylation [MYR] and S-palmitoylation [PAL]) are crucial modifications affecting 2 to 4% of eukaryotic proteins. The role of these modifications is to target proteins to membranes. Predictive tools have revealed unexpected targets of these acylations in Arabidopsis thaliana and other plants. However, little is known about how N-terminal lipidation governs membrane compartmentalization of proteins in plants. We show here that h-type thioredoxins (h-TRXs) cluster in four evolutionary subgroups displaying strictly conserved N-terminal modifications. It was predicted that one subgroup undergoes only MYR and another undergoes both MYR and PAL. We used plant TRXs as a model protein family to explore the effect of MYR alone or MYR and PAL in the same family of proteins. We used a high-throughput biochemical strategy to assess MYR of specific TRXs. Moreover, various TRX-green fluorescent protein fusions revealed that MYR localized protein to the endomembrane system and that partitioning between this membrane compartment and the cytosol correlated with the catalytic efficiency of the N-myristoyltransferase acting at the N terminus of the TRXs. Generalization of these results was obtained using several randomly selected Arabidopsis proteins displaying a MYR site only. Finally, we demonstrated that a palmitoylatable Cys residue flanking the MYR site is crucial to localize proteins to micropatching zones of the plasma membrane.
Figures

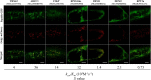
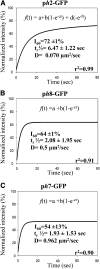
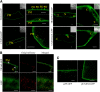


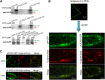
References
-
- Adjobo-Hermans M.J., Goedhart J., Gadella T.W., Jr (2006). Plant G protein heterotrimers require dual lipidation motifs of Galpha and Ggamma and do not dissociate upon activation. J. Cell Sci. 119: 5087–5097 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

