Comparative proteomics of chloroplasts envelopes from bundle sheath and mesophyll chloroplasts reveals novel membrane proteins with a possible role in c4-related metabolite fluxes and development
- PMID: 23543921
- PMCID: PMC3610082
- DOI: 10.3389/fpls.2013.00065
Comparative proteomics of chloroplasts envelopes from bundle sheath and mesophyll chloroplasts reveals novel membrane proteins with a possible role in c4-related metabolite fluxes and development
Abstract
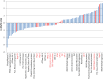
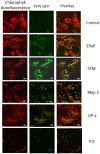
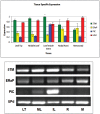
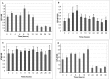
As the world population grows, our need for food increases drastically. Limited amounts of arable land lead to a competition between food and fuel crops, while changes in the global climate may impact future crop yields. Thus, a second "green revolution" will need a better understanding of the processes essential for plant growth and development. One approach toward the solution of this problem is to better understand regulatory and transport processes in C4 plants. C4 plants display an up to 10-fold higher apparent CO2 assimilation and higher yields while maintaining high water use efficiency. This requires differential regulation of mesophyll (M) and bundle sheath (BS) chloroplast development as well as higher metabolic fluxes of photosynthetic intermediates between cells and particularly across chloroplast envelopes. While previous analyses of overall chloroplast membranes have yielded significant insight, our comparative proteomics approach using enriched BS and M chloroplast envelopes of Zea mays allowed us to identify 37 proteins of unknown function that have not been seen in these earlier studies. We identified 280 proteins, 84% of which are known/predicted to be present in chloroplasts. Seventy-four percent have a known or predicted membrane association. Twenty-one membrane proteins were 2-15 times more abundant in BS cells, while 36 of the proteins were more abundant in M chloroplast envelopes. These proteins could represent additional candidates of proteins essential for development or metabolite transport processes in C4 plants. RT-PCR confirmed differential expression of 13 candidate genes. Chloroplast association for seven proteins was confirmed using YFP/GFP labeling. Gene expression of four putative transporters was examined throughout the leaf and during the greening of leaves. Genes for a PIC-like protein and an ER-AP-like protein show an early transient increase in gene expression during the transition to light. In addition, PIC gene expression is increased in the immature part of the leaf and was lower in the fully developed parts of the leaf, suggesting a need for/incorporation of the protein during chloroplast development.
Keywords: C4 plant; bundle sheath cells; chloroplast envelope proteins; mesophyll cells; photosynthesis.
Figures


Similar articles
-
Comparative proteomics of chloroplast envelopes from C3 and C4 plants reveals specific adaptations of the plastid envelope to C4 photosynthesis and candidate proteins required for maintaining C4 metabolite fluxes.Plant Physiol. 2008 Sep;148(1):568-79. doi: 10.1104/pp.108.121012. Epub 2008 Jul 3. Plant Physiol. 2008. PMID: 18599648 Free PMC article.
-
A novel RNA binding protein affects rbcL gene expression and is specific to bundle sheath chloroplasts in C4 plants.BMC Plant Biol. 2013 Sep 22;13:138. doi: 10.1186/1471-2229-13-138. BMC Plant Biol. 2013. PMID: 24053212 Free PMC article.
-
The role of chloroplast movement in C4 photosynthesis: a theoretical analysis using a three-dimensional reaction-diffusion model for maize.J Exp Bot. 2023 Aug 3;74(14):4125-4142. doi: 10.1093/jxb/erad138. J Exp Bot. 2023. PMID: 37083863 Free PMC article.
-
Cell-type-specific differentiation of chloroplasts in C4 plants.Trends Plant Sci. 2009 Feb;14(2):100-9. doi: 10.1016/j.tplants.2008.11.006. Epub 2009 Jan 21. Trends Plant Sci. 2009. PMID: 19162526 Review.
-
C4 rice engineering, beyond installing a C4 cycle.Plant Physiol Biochem. 2024 Jan;206:108256. doi: 10.1016/j.plaphy.2023.108256. Epub 2023 Dec 7. Plant Physiol Biochem. 2024. PMID: 38091938 Review.
Cited by
-
Unknown components of the plastidial permeome.Front Plant Sci. 2014 Aug 19;5:410. doi: 10.3389/fpls.2014.00410. eCollection 2014. Front Plant Sci. 2014. PMID: 25191333 Free PMC article. Review.
-
The Differences between NAD-ME and NADP-ME Subtypes of C4 Photosynthesis: More than Decarboxylating Enzymes.Front Plant Sci. 2016 Oct 13;7:1525. doi: 10.3389/fpls.2016.01525. eCollection 2016. Front Plant Sci. 2016. PMID: 27790235 Free PMC article. Review.
-
iTRAQ-based analysis of developmental dynamics in the soybean leaf proteome reveals pathways associated with leaf photosynthetic rate.Mol Genet Genomics. 2016 Aug;291(4):1595-605. doi: 10.1007/s00438-016-1202-3. Epub 2016 Apr 5. Mol Genet Genomics. 2016. PMID: 27048574
-
Application of proteomics for improving crop protection/artificial regulation.Front Plant Sci. 2013 Dec 19;4:522. doi: 10.3389/fpls.2013.00522. eCollection 2013. Front Plant Sci. 2013. PMID: 24391656 Free PMC article. No abstract available.
-
Comparative proteomic analysis of a membrane-enriched fraction from flag leaves reveals responses to chemical hybridization agent SQ-1 in wheat.Front Plant Sci. 2015 Aug 26;6:669. doi: 10.3389/fpls.2015.00669. eCollection 2015. Front Plant Sci. 2015. PMID: 26379693 Free PMC article.
References
-
- Bassham J., Benson A., Calvin M. (1950). The path of carbon in photosynthesis. J. Biol. Chem. 185, 781–787 - PubMed
-
- Bräutigam A., Hoffmann-Benning S., Weber A. P. M. (2008). Comparative proteomics of chloroplast envelopes from C3 and C4 plants reveals specific adaptations of the plastid envelope to C4 photosynthesis and candidate proteins required for maintaining C4 metabolite fluxes. Plant Physiol. 148, 568–57910.1104/pp.108.121012 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

