Replication clamps and clamp loaders
- PMID: 23545418
- PMCID: PMC3683903
- DOI: 10.1101/cshperspect.a010165
Replication clamps and clamp loaders
Abstract
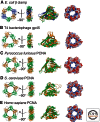
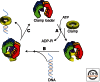
To achieve the high degree of processivity required for DNA replication, DNA polymerases associate with ring-shaped sliding clamps that encircle the template DNA and slide freely along it. The closed circular structure of sliding clamps necessitates an enzyme-catalyzed mechanism, which not only opens them for assembly and closes them around DNA, but specifically targets them to sites where DNA synthesis is initiated and orients them correctly for replication. Such a feat is performed by multisubunit complexes known as clamp loaders, which use ATP to open sliding clamp rings and place them around the 3' end of primer-template (PT) junctions. Here we discuss the structure and composition of sliding clamps and clamp loaders from the three domains of life as well as T4 bacteriophage, and provide our current understanding of the clamp-loading process.
Figures



References
-
- Ahmadian MR, Stege P, Scheffzek K, Wittinghofer A 1997. Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat Struct Biol 4: 686–689 - PubMed
-
- Alley SC, Jones AD, Soumillion P, Benkovic SJ 1999a. The carboxyl terminus of the bacteriophage T4 DNA polymerase contacts its sliding clamp at the subunit interface. J Biol Chem 274: 24485–24489 - PubMed
-
- Alley SC, Shier VK, Abel-Santos E, Sexton DJ, Soumillion P, Benkovic SJ 1999b. Sliding clamp of the bacteriophage T4 polymerase has open and closed subunit interfaces in solution. Biochemistry 38: 7696–7709 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
