Endoplasmic reticulum structure and interconnections with other organelles
- PMID: 23545422
- PMCID: PMC3683900
- DOI: 10.1101/cshperspect.a013227
Endoplasmic reticulum structure and interconnections with other organelles
Abstract
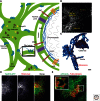
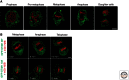
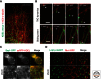
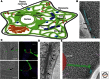
The endoplasmic reticulum (ER) is a large, continuous membrane-bound organelle comprised of functionally and structurally distinct domains including the nuclear envelope, peripheral tubular ER, peripheral cisternae, and numerous membrane contact sites at the plasma membrane, mitochondria, Golgi, endosomes, and peroxisomes. These domains are required for multiple cellular processes, including synthesis of proteins and lipids, calcium level regulation, and exchange of macromolecules with various organelles at ER-membrane contact sites. The ER maintains its unique overall structure regardless of dynamics or transfer at ER-organelle contacts. In this review, we describe the numerous factors that contribute to the structure of the ER.
Figures
References
-
- Achleitner G, Gaigg B, Krasser A, Kainersdorfer E, Kohlwein SD, Perktold A, Zellnig G, Daum G 1999. Association between the endoplasmic reticulum and mitochondria of yeast facilitates interorganelle transport of phospholipids through membrane contact. Eur J Biochem 264: 545–553 - PubMed
-
- Anderson DJ, Hetzer MW 2007. Nuclear envelope formation by chromatin-mediated reorganization of the endoplasmic reticulum. Nat Cell Biol 9: 1160–1166 - PubMed
-
- Anderson DJ, Hetzer MW 2008b. Shaping the endoplasmic reticulum into the nuclear envelope. J Cell Sci 121: 137–142 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
