Structure of an atypical FeoB G-domain reveals a putative domain-swapped dimer
- PMID: 23545645
- PMCID: PMC3614164
- DOI: 10.1107/S1744309113005939
Structure of an atypical FeoB G-domain reveals a putative domain-swapped dimer
Abstract
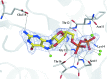
FeoB is a transmembrane protein involved in ferrous iron uptake in prokaryotic organisms. FeoB comprises a cytoplasmic soluble domain termed NFeoB and a C-terminal polytopic transmembrane domain. Recent structures of NFeoB have revealed two structural subdomains: a canonical GTPase domain and a five-helix helical domain. The GTPase domain hydrolyses GTP to GDP through a well characterized mechanism, a process which is required for Fe(2+) transport. In contrast, the precise role of the helical domain has not yet been fully determined. Here, the structure of the cytoplasmic domain of FeoB from Gallionella capsiferriformans is reported. Unlike recent structures of NFeoB, the G. capsiferriformans NFeoB structure is highly unusual in that it does not contain a helical domain. The crystal structures of both apo and GDP-bound protein forms a domain-swapped dimer.
Keywords: FeoB; Gallionella capsiferriformans; NFeoB; domain-swapped dimer.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

