A novel knowledge representation framework for the statistical validation of quantitative imaging biomarkers
- PMID: 23546775
- PMCID: PMC3705009
- DOI: 10.1007/s10278-013-9598-3
A novel knowledge representation framework for the statistical validation of quantitative imaging biomarkers
Abstract
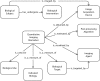
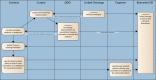
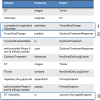
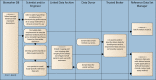
Quantitative imaging biomarkers are of particular interest in drug development for their potential to accelerate the drug development pipeline. The lack of consensus methods and carefully characterized performance hampers the widespread availability of these quantitative measures. A framework to support collaborative work on quantitative imaging biomarkers would entail advanced statistical techniques, the development of controlled vocabularies, and a service-oriented architecture for processing large image archives. Until now, this framework has not been developed. With the availability of tools for automatic ontology-based annotation of datasets, coupled with image archives, and a means for batch selection and processing of image and clinical data, imaging will go through a similar increase in capability analogous to what advanced genetic profiling techniques have brought to molecular biology. We report on our current progress on developing an informatics infrastructure to store, query, and retrieve imaging biomarker data across a wide range of resources in a semantically meaningful way that facilitates the collaborative development and validation of potential imaging biomarkers by many stakeholders. Specifically, we describe the semantic components of our system, QI-Bench, that are used to specify and support experimental activities for statistical validation in quantitative imaging.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

