Pathway-based personalized analysis of cancer
- PMID: 23547110
- PMCID: PMC3631698
- DOI: 10.1073/pnas.1219651110
Pathway-based personalized analysis of cancer
Abstract
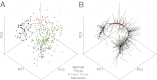
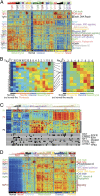
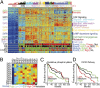
We introduce Pathifier, an algorithm that infers pathway deregulation scores for each tumor sample on the basis of expression data. This score is determined, in a context-specific manner, for every particular dataset and type of cancer that is being investigated. The algorithm transforms gene-level information into pathway-level information, generating a compact and biologically relevant representation of each sample. We demonstrate the algorithm's performance on three colorectal cancer datasets and two glioblastoma multiforme datasets and show that our multipathway-based representation is reproducible, preserves much of the original information, and allows inference of complex biologically significant information. We discovered several pathways that were significantly associated with survival of glioblastoma patients and two whose scores are predictive of survival in colorectal cancer: CXCR3-mediated signaling and oxidative phosphorylation. We also identified a subclass of proneural and neural glioblastoma with significantly better survival, and an EGF receptor-deregulated subclass of colon cancers.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
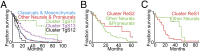
References
-
- Thomas DC, et al. American Association for Cancer Research Approaches to complex pathways in molecular epidemiology: Summary of a special conference of the American Association for Cancer Research. Cancer Res. 2008;68(24):10028–10030. - PubMed
-
- Segal E, Friedman N, Koller D, Regev A. A module map showing conditional activity of expression modules in cancer. Nat Genet. 2004;36(10):1090–1098. - PubMed
-
- Bild AH, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439(7074):353–357. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

